Localizing recent adaptive evolution in the human genome
- PMID: 17542651
- PMCID: PMC1885279
- DOI: 10.1371/journal.pgen.0030090
Localizing recent adaptive evolution in the human genome
Abstract
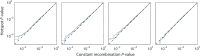
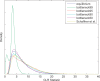
Identifying genomic locations that have experienced selective sweeps is an important first step toward understanding the molecular basis of adaptive evolution. Using statistical methods that account for the confounding effects of population demography, recombination rate variation, and single-nucleotide polymorphism ascertainment, while also providing fine-scale estimates of the position of the selected site, we analyzed a genomic dataset of 1.2 million human single-nucleotide polymorphisms genotyped in African-American, European-American, and Chinese samples. We identify 101 regions of the human genome with very strong evidence (p < 10(-5)) of a recent selective sweep and where our estimate of the position of the selective sweep falls within 100 kb of a known gene. Within these regions, genes of biological interest include genes in pigmentation pathways, components of the dystrophin protein complex, clusters of olfactory receptors, genes involved in nervous system development and function, immune system genes, and heat shock genes. We also observe consistent evidence of selective sweeps in centromeric regions. In general, we find that recent adaptation is strikingly pervasive in the human genome, with as much as 10% of the genome affected by linkage to a selective sweep.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures




References
-
- Smith NGC, Eyre-Walker A. Adaptive protein evolution in Drosophila . Nature. 2002;415:1022–1024. - PubMed
-
- Clark AG, Glanowski S, Nielsen R, Thomas P, Kejariwal A, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. - PubMed
-
- Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. - PubMed

