Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network
- PMID: 17543864
- PMCID: PMC2705290
- DOI: 10.1016/j.devcel.2007.04.002
Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network
Abstract
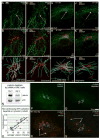
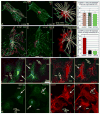
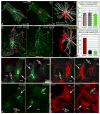
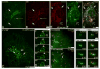
Proper organization of microtubule arrays is essential for intracellular trafficking and cell motility. It is generally assumed that most if not all microtubules in vertebrate somatic cells are formed by the centrosome. Here we demonstrate that a large number of microtubules in untreated human cells originate from the Golgi apparatus in a centrosome-independent manner. Both centrosomal and Golgi-emanating microtubules need gamma-tubulin for nucleation. Additionally, formation of microtubules at the Golgi requires CLASPs, microtubule-binding proteins that selectively coat noncentrosomal microtubule seeds. We show that CLASPs are recruited to the trans-Golgi network (TGN) at the Golgi periphery by the TGN protein GCC185. In sharp contrast to radial centrosomal arrays, microtubules nucleated at the peripheral Golgi compartment are preferentially oriented toward the leading edge in motile cells. We propose that Golgi-emanating microtubules contribute to the asymmetric microtubule networks in polarized cells and support diverse processes including post-Golgi transport to the cell front.
Figures



Comment in
-
CLASPing microtubules at the trans-Golgi network.Dev Cell. 2007 Jun;12(6):839-40. doi: 10.1016/j.devcel.2007.05.007. Dev Cell. 2007. PMID: 17543853
References
-
- Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, Galjart N. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. - PubMed
-
- Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155–4163. - PubMed
-
- Bugnard E, Zaal KJ, Ralston E. Reorganization of microtubule nucleation during muscle differentiation. Cell Motil Cytoskeleton. 2005;60:1–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

