Efficient and exclusive induction of Tet repressor by the oligopeptide Tip results from co-variation of their interaction site
- PMID: 17545198
- PMCID: PMC1919500
- DOI: 10.1093/nar/gkm357
Efficient and exclusive induction of Tet repressor by the oligopeptide Tip results from co-variation of their interaction site
Abstract
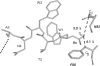
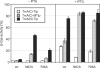
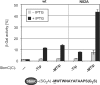
Protein-protein interactions are an important element of signal transfer within and between organisms. They are mainly mediated by short oligopeptide motifs and represent a widely used alternative to small, organic molecules for conveying information. The transcription factor TetR, a regulator of tetracycline resistance in Gram-negative bacteria, is naturally induced by tetracycline or its derivatives. The oligopeptide Tip (Transcription inducing peptide) fused to either N- or C-terminus of Thioredoxin A (TrxA) has been isolated as an artificial inducer for TetR in Escherichia coli. This inducing property can be exploited to monitor the in vivo expression of a protein of interest by fusing Tip to its C-terminus. We improve the induction efficiency of Tip by adding an aromatic amino acid before residue 1 of Tip in C-terminal fusions to TrxA. The induction efficiency of that modified TrxA-Tip fusion is further enhanced when the effector-binding pocket of TetR is enlarged by the N82A and F86A mutations. The double mutant is also insensitive to induction by tetracyclines. Thus, Tip is an exclusive inducer of this TetR variant, representing the first example of fully converting a small molecular weight effector-dependent transcription factor into one depending solely on protein-protein recognition.
Figures

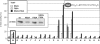

Similar articles
-
Functionally important residues of the Tet repressor inducing peptide TIP determined by a complete mutational analysis.Gene. 2008 Nov 1;423(2):201-6. doi: 10.1016/j.gene.2008.07.004. Epub 2008 Jul 12. Gene. 2008. PMID: 18672042
-
How an agonist peptide mimics the antibiotic tetracycline to induce Tet-repressor.J Mol Biol. 2007 May 4;368(3):780-90. doi: 10.1016/j.jmb.2007.02.030. Epub 2007 Feb 22. J Mol Biol. 2007. PMID: 17374541
-
Conformational changes of the Tet repressor induced by tetracycline trapping.J Mol Biol. 1998 Jun 5;279(2):439-47. doi: 10.1006/jmbi.1998.1775. J Mol Biol. 1998. PMID: 9642048
-
Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes.Eur J Biochem. 2003 Aug;270(15):3109-21. doi: 10.1046/j.1432-1033.2003.03694.x. Eur J Biochem. 2003. PMID: 12869186 Review.
-
Gene regulation by tetracyclines.Genet Eng (N Y). 2004;26:255-77. doi: 10.1007/978-0-306-48573-2_13. Genet Eng (N Y). 2004. PMID: 15387301 Review.
Cited by
-
Noise-reducing optogenetic negative-feedback gene circuits in human cells.Nucleic Acids Res. 2019 Aug 22;47(14):7703-7714. doi: 10.1093/nar/gkz556. Nucleic Acids Res. 2019. PMID: 31269201 Free PMC article.
-
A novel TetR-regulating peptide turns off rtTA-mediated activation of gene expression.PLoS One. 2014 May 8;9(5):e96546. doi: 10.1371/journal.pone.0096546. eCollection 2014. PLoS One. 2014. PMID: 24810590 Free PMC article.
-
Status quo of tet regulation in bacteria.Microb Biotechnol. 2022 Apr;15(4):1101-1119. doi: 10.1111/1751-7915.13926. Epub 2021 Oct 29. Microb Biotechnol. 2022. PMID: 34713957 Free PMC article. Review.
-
Intracellular monitoring of target protein production in Staphylococcus aureus by peptide tag-induced reporter fluorescence.Microb Biotechnol. 2012 Jan;5(1):129-34. doi: 10.1111/j.1751-7915.2011.00304.x. Epub 2011 Sep 29. Microb Biotechnol. 2012. PMID: 21958360 Free PMC article.
-
Tetracycline-tet repressor binding specificity: insights from experiments and simulations.Biophys J. 2009 Nov 18;97(10):2829-38. doi: 10.1016/j.bpj.2009.08.050. Biophys J. 2009. PMID: 19917238 Free PMC article.
References
-
- Ogawa H, Qiu Y, Ogata CM, Misono KS. Crystal structure of hormone-bound atrial natriuretic peptide receptor extracellular domain: rotation mechanism for transmembrane signal transduction. J. Biol. Chem. 2004;279:28625–28631. - PubMed
-
- Rosenfeld Y, Papo N, Shai Y. Endotoxin (lipopolysaccharide) neutralization by innate immunity host-defense peptides. Peptide properties and plausible modes of action. J. Biol. Chem. 2006;281:1636–1643. - PubMed
-
- Mourez M, Kane RS, Mogridge J, Metallo S, Deschatelets P, Sellman BR, Whitesides GM, Collier RJ. Designing a polyvalent inhibitor of anthrax toxin. Nat. Biotechnol. 2001;19:958–961. - PubMed
-
- Desjobert C, de Soultrait VR, Faure A, Parissi V, Litvak S, Tarrago-Litvak L, Fournier M. Identification by phage display selection of a short peptide able to inhibit only the strand transfer reaction catalyzed by human immunodeficiency virus type 1 integrase. Biochemistry. 2004;43:13097–13105. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

