FLAN: a web server for influenza virus genome annotation
- PMID: 17545199
- PMCID: PMC1933127
- DOI: 10.1093/nar/gkm354
FLAN: a web server for influenza virus genome annotation
Abstract
FLAN (short for FLu ANnotation), the NCBI web server for genome annotation of influenza virus (http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/annotation.cgi) is a tool for user-provided influenza A virus or influenza B virus sequences. It can validate and predict protein sequences encoded by an input flu sequence. The input sequence is BLASTed against a database containing influenza sequences to determine the virus type (A or B), segment (1 through 8) and subtype for the hemagglutinin and neuraminidase segments of influenza A virus. For each segment/subtype of the viruses, a set of sample protein sequences is maintained. The input sequence is then aligned against the corresponding protein set with a 'Protein to nucleotide alignment tool' (ProSplign). The translated product from the best alignment to the sample protein sequence is used as the predicted protein encoded by the input sequence. The output can be a feature table that can be used for sequence submission to GenBank (by Sequin or tbl2asn), a GenBank flat file, or the predicted protein sequences in FASTA format. A message showing the length of the input sequence, the predicted virus type, segment and subtype for the hemagglutinin and neuraminidase segments of Influenza A virus will also be displayed.
Figures

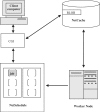
References
-
- Ghedin E, Sengamalay NA, Shumway M, Zaborsky J, Feldblyum T, Subbu V, Spiro DJ, Sitz J, Koo H, et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature. 2005;437:1162–1166. - PubMed
-
- Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, et al. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–1580. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed

