Folding and unfolding of gammaTIM monomers and dimers
- PMID: 17545246
- PMCID: PMC1965449
- DOI: 10.1529/biophysj.107.108068
Folding and unfolding of gammaTIM monomers and dimers
Abstract
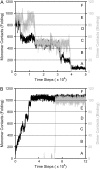
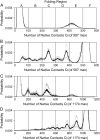
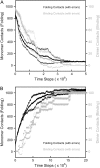
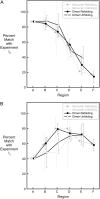
Kinetic simulations of the folding and unfolding of triosephosphate isomerase (TIM) from yeast were conducted using a single monomer gammaTIM polypeptide chain that folds as a monomer and two gammaTIM chains that fold to the native dimer structure. The basic protein model used was a minimalist Gō model using the native structure to determine attractive energies in the protein chain. For each simulation type--monomer unfolding, monomer refolding, dimer unfolding, and dimer refolding--thirty simulations were conducted, successfully capturing each reaction in full. Analysis of the simulations demonstrates four main conclusions. First, all four simulation types have a similar "folding order", i.e., they have similar structures in intermediate stages of folding between the unfolded and folded state. Second, despite this similarity, different intermediate stages are more or less populated in the four different simulations, with 1), no intermediates populated in monomer unfolding; 2), two intermediates populated with beta(2)-beta(4) and beta(1)-beta(5) regions folded in monomer refolding; 3), two intermediates populated with beta(2)-beta(3) and beta(2)-beta(4) regions folded in dimer unfolding; and 4), two intermediates populated with beta(1)-beta(5) and beta(1)-beta(5) + beta(6) + beta(7) + beta(8) regions folded in dimer refolding. Third, simulations demonstrate that dimer binding and unbinding can occur early in the folding process before complete monomer-chain folding. Fourth, excellent agreement is found between the simulations and MPAX (misincorporation proton alkyl exchange) experiments. In total, this agreement demonstrates that the computational Gō model is accurate for gammaTIM and that the energy landscape of gammaTIM appears funneled to the native state.
Figures

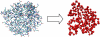
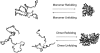


References
-
- Onuchic, J. N., Z. Luthey-Schulten, and P. G. Wolynes. 1997. Theory of protein folding: the energy landscape perspective. Annu. Rev. Phys. Chem. 48:545–600. - PubMed
-
- Garel, T., and H. Orland. 1988. Mean-field model for protein folding. Europhys. Lett. (Switzerland). 6:307–310.
-
- Shakhnovich, E. I., and A. M. Gutin. 1989. The nonergodic (spin-glass-like) phase of heteropolymer with quenched disordered sequence of links. Europhys. Lett. (Switzerland). 8:327–332.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

