Crystal structures of the receiver domain of the response regulator PhoP from Escherichia coli in the absence and presence of the phosphoryl analog beryllofluoride
- PMID: 17545283
- PMCID: PMC1952025
- DOI: 10.1128/JB.00049-07
Crystal structures of the receiver domain of the response regulator PhoP from Escherichia coli in the absence and presence of the phosphoryl analog beryllofluoride
Abstract
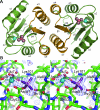
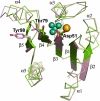
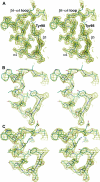
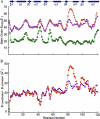
The response regulator PhoP is part of the PhoQ/PhoP two-component system involved in responses to depletion of extracellular Mg(2+). Here, we report the crystal structures of the receiver domain of Escherichia coli PhoP determined in the absence and presence of the phosphoryl analog beryllofluoride. In the presence of beryllofluoride, the active receiver domain forms a twofold symmetric dimer similar to that seen in structures of other regulatory domains from the OmpR/PhoB family, providing further evidence that members of this family utilize a common mode of dimerization in the active state. In the absence of activating agents, the PhoP receiver domain crystallizes with a similar structure, consistent with the previous observation that high concentrations can promote an active state of PhoP independent of phosphorylation.
Figures

References
-
- Bijlsma, J. J., and E. A. Groisman. 2005. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol. 57:85-96. - PubMed
-
- Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

