Human Polycomb group EED protein negatively affects HIV-1 assembly and release
- PMID: 17547741
- PMCID: PMC1899515
- DOI: 10.1186/1742-4690-4-37
Human Polycomb group EED protein negatively affects HIV-1 assembly and release
Abstract
Background: The human EED protein, a member of the superfamily of Polycomb group (PcG) proteins with WD-40 repeats, has been found to interact with three HIV-1 components, namely the structural Gag matrix protein (MA), the integrase enzyme (IN) and the Nef protein. The aim of the present study was to analyze the possible biological role of EED in HIV-1 replication, using the HIV-1-based vector HIV-Luc and EED protein expressed by DNA transfection of 293T cells.
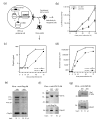
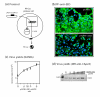
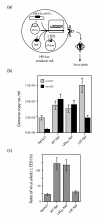
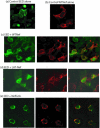
Results: During the early phase of HIV-1 infection, a slight negative effect on virus infectivity occurred in EED-expressing cells, which appeared to be dependent on EED-MA interaction. At late times post infection, EED caused an important reduction of virus production, from 20- to 25-fold as determined by CAp24 immunoassay, to 10- to 80-fold based on genomic RNA levels, and this decrease was not due to a reduction of Gag protein synthesis. Coexpression of WTNef, or the non-N-myristoylated mutant NefG2A, restored virus yields to levels obtained in the absence of exogenous EED protein. This effect was not observed with mutant NefDelta57 mimicking the Nef core, or with the lipid raft-retargeted fusion protein LAT-Nef. LATAA-Nef, a mutant defective in the lipid raft addressing function, had the same anti-EED effect as WTNef. Cell fractionation and confocal imaging showed that, in the absence of Nef, EED mainly localized in membrane domains different from the lipid rafts. Upon co-expression with WTNef, NefG2A or LATAA-Nef, but not with NefDelta57 or LAT-Nef, EED was found to relocate into an insoluble fraction along with Nef protein. Electron microscopy of HIV-Luc producer cells overexpressing EED showed significant less virus budding at the cell surface compared to control cells, and ectopic assembly and clustering of nuclear pore complexes within the cytoplasm.
Conclusion: Our data suggested that EED exerted an antiviral activity at the late stage of HIV-1 replication, which included genomic RNA packaging and virus assembly, resulting possibly from a mistrafficking of viral genomic RNA (gRNA) or gRNA/Gag complex. Nef reversed the EED negative effect on virus production, a function which required the integrity of the Nef N-terminal domain, but not its N-myristoyl group. The antagonistic effect of Nef correlated with a cellular redistribution of both EED and Nef.
Figures






References
-
- Sewalt RGAB, van der Vlag J, Gunster MJ, Hamer KM, den Blaauwen JL, Satijn DPE, Hendrix T, van Driel R, Otte AP. Characterization of interactions between the mammalian Polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian Polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–3595. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

