Prion protein repeat expansion results in increased aggregation and reveals phenotypic variability
- PMID: 17548473
- PMCID: PMC1952097
- DOI: 10.1128/MCB.02127-06
Prion protein repeat expansion results in increased aggregation and reveals phenotypic variability
Abstract
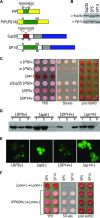
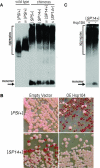
Mammalian prion diseases are fatal neurodegenerative disorders dependent on the prion protein PrP. Expansion of the oligopeptide repeats (ORE) found in PrP is associated with inherited prion diseases. Patients with ORE frequently harbor PrP aggregates, but other factors may contribute to pathology, as they often present with unexplained phenotypic variability. We created chimeric yeast-mammalian prion proteins to examine the influence of the PrP ORE on prion properties in yeast. Remarkably, all chimeric proteins maintained prion characteristics. The largest repeat expansion chimera displayed a higher propensity to maintain a self-propagating aggregated state. Strikingly, the repeat expansion conferred increased conformational flexibility, as observed by enhanced phenotypic variation. Furthermore, the repeat expansion chimera displayed an increased rate of prion conversion, but only in the presence of another aggregate, the [RNQ+] prion. We suggest that the PrP ORE increases the conformational flexibility of the prion protein, thereby enhancing the formation of multiple distinct aggregate structures and allowing more frequent prion conversion. Both of these characteristics may contribute to the phenotypic variability associated with PrP repeat expansion diseases.
Figures



References
-
- Bagriantsev, S., and S. W. Liebman. 2004. Specificity of prion assembly in vivo. J. Biol. Chem. 279:51042-51048. - PubMed
-
- Bessen, R. A., D. A. Kocisko, G. J. Raymond, S. Nandan, P. T. Lansbury, and B. Caughey. 1995. Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature 375:698-700. - PubMed
-
- Bessen, R. A., and R. F. Marsh. 1992. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J. Gen. Virol. 73(Pt. 2):329-334. - PubMed
-
- Borchsenius, A. S., S. Muller, G. P. Newnam, S. G. Inge-Vechtomov, and Y. O. Chernoff. 2006. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr. Genet. 49:21-29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
