New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate
- PMID: 17548499
- PMCID: PMC2043241
- DOI: 10.1128/AAC.00339-07
New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate
Abstract
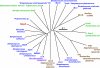
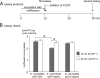
Plasmid-mediated Qnr and AAC(6')-Ib-cr have been recognized as new molecular mechanisms affecting fluoroquinolone (FQ) resistance. C316, an Escherichia coli strain demonstrating resistance to various FQs, was isolated in Japan. Resistance to FQs was augmented in an E. coli CSH2 transconjugant, but PCR failed to detect qnr genes, suggesting the presence of novel plasmid-mediated FQ resistance mechanisms. Susceptibility tests, DNA manipulation, and analyses of the gene and its product were performed to characterize the genetic determinant. A novel FQ-resistant gene, qepA, was identified in a plasmid, pHPA, of E. coli C316, and both qepA and rmtB genes were mediated by a probable transposable element flanked by two copies of IS26. Levels of resistance to norfloxacin, ciprofloxacin, and enrofloxacin were significantly elevated in E. coli transformants harboring qepA under AcrB-TolC-deficient conditions. QepA showed considerable similarities to transporters belonging to the 14-transmembrane-segment family of environmental actinomycetes. The effect of carbonyl cyanide m-chlorophenylhydrazone (CCCP) on accumulation of norfloxacin was assayed in a qepA-harboring E. coli transformant. The intracellular accumulation of norfloxacin was decreased in a qepA-expressing E. coli transformant, but this phenomenon was canceled by CCCP. The augmented FQ resistance level acquired by the probable intergeneric transfer of a gene encoding a major facilitator superfamily-type efflux pump from some environmental microbes to E. coli was first identified. Surveillance of the qepA-harboring clinical isolates should be encouraged to minimize further dissemination of the kind of plasmid-dependent FQ resistance determinants among pathogenic microbes.
Figures



References
-
- Bogaerts, P., M. Galimand, C. Bauraing, A. Deplano, R. Vanhoof, R. De Mendonca, H. Rodriguez-Villalobos, M. Struelens, and Y. Glupczynski. 2007. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J. Antimicrob. Chemother. 59:459-464. - PubMed
-
- Chen, L., Z. L. Chen, J. H. Liu, Z. L. Zeng, J. Y. Ma, and H. X. Jiang. 2007. Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J. Antimicrob. Chemother. 59:880-885. - PubMed
-
- Doi, Y., and Y. Arakawa. 2007. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 45:88-94. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

