Stochastic innovation as a mechanism by which catalysts might self-assemble into chemical reaction networks
- PMID: 17548812
- PMCID: PMC1891208
- DOI: 10.1073/pnas.0703522104
Stochastic innovation as a mechanism by which catalysts might self-assemble into chemical reaction networks
Abstract
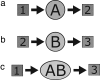
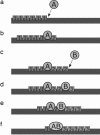
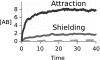
We develop a computer model for how two different chemical catalysts in solution, A and B, could be driven to form AB complexes, based on the concentration gradients of a substrate or product that they share in common. If A's product is B's substrate, B will be attracted to A, mediated by a common resource that is not otherwise plentiful in the environment. By this simple physicochemical mechanism, chemical reactions could spontaneously associate to become chained together in solution. According to the model, such catalyst self-association processes may resemble other processes of "stochastic innovation," such as Darwinian evolution in biology, that involve a search among options, a selection among those options, and then a lock-in of that selection. Like Darwinian processes, this simple chemical process exhibits cooperation, competition, innovation, and a preference for consistency. This model may be useful for understanding organizational processes in prebiotic chemistry and for developing new kinds of self-organization in chemically reacting systems.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Discreteness-induced slow relaxation in reversible catalytic reaction networks.Phys Rev E Stat Nonlin Soft Matter Phys. 2010 May;81(5 Pt 1):051920. doi: 10.1103/PhysRevE.81.051920. Epub 2010 May 21. Phys Rev E Stat Nonlin Soft Matter Phys. 2010. PMID: 20866274
-
Molecular mechanics in biology: from structure to function, taking account of solvation.Annu Rev Biophys Biomol Struct. 1994;23:847-63. doi: 10.1146/annurev.bb.23.060194.004215. Annu Rev Biophys Biomol Struct. 1994. PMID: 7919800 Review. No abstract available.
-
Time-ordered product expansions for computational stochastic system biology.Phys Biol. 2013 Jun;10(3):035009. doi: 10.1088/1478-3975/10/3/035009. Epub 2013 Jun 4. Phys Biol. 2013. PMID: 23735739 Free PMC article.
-
The origin and spread of a cooperative replicase in a prebiotic chemical system.J Theor Biol. 2015 Jan 7;364:249-59. doi: 10.1016/j.jtbi.2014.09.019. Epub 2014 Sep 22. J Theor Biol. 2015. PMID: 25245369
-
Ribose Selected as Precursor to Life.DNA Cell Biol. 2020 Feb;39(2):177-186. doi: 10.1089/dna.2019.4943. Epub 2019 Dec 4. DNA Cell Biol. 2020. PMID: 31804855 Review.
Cited by
-
Compositional inheritance: comparison of self-assembly and catalysis.Orig Life Evol Biosph. 2008 Oct;38(5):399-418. doi: 10.1007/s11084-008-9143-4. Epub 2008 Jul 18. Orig Life Evol Biosph. 2008. PMID: 18636340
-
Driving forces in the origins of life.Open Biol. 2021 Feb;11(2):200324. doi: 10.1098/rsob.200324. Epub 2021 Feb 3. Open Biol. 2021. PMID: 33529553 Free PMC article.
-
The last universal common ancestor: emergence, constitution and genetic legacy of an elusive forerunner.Biol Direct. 2008 Jul 9;3:29. doi: 10.1186/1745-6150-3-29. Biol Direct. 2008. PMID: 18613974 Free PMC article. Review.
-
Nanoscale Catalyst Chemotaxis Can Drive the Assembly of Functional Pathways.J Phys Chem B. 2021 Aug 12;125(31):8781-8786. doi: 10.1021/acs.jpcb.1c04498. Epub 2021 Jul 29. J Phys Chem B. 2021. PMID: 34324352 Free PMC article.
References
-
- Mayr E. What Evolution Is. New York: Basic Books; 2001.
-
- Hebb DO. The Organization of Behavior. New York: Wiley; 1949.
-
- Goda Y, Davis GW. Neuron. 2003;40:243–264. - PubMed
-
- Alberts B, Johnson A, Lewis J, Raff M., Roberts K, Walter P. Mol Biol Cell. New York: Garland Science; 2002. p. 1238.

