nanos gene control DNA mediates developmentally regulated transposition in the yellow fever mosquito Aedes aegypti
- PMID: 17548819
- PMCID: PMC1891237
- DOI: 10.1073/pnas.0701515104
nanos gene control DNA mediates developmentally regulated transposition in the yellow fever mosquito Aedes aegypti
Abstract
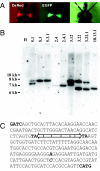
Transposable elements (TEs) are proposed as a basis for developing drive systems to spread pathogen resistance genes through vector mosquito populations. The use of transcriptional and translational control DNA elements from genes expressed specifically in the insect germ line to mediate transposition offers possibilities for mitigating some of the concerns about transgene behavior in the target vector species and eliminating effects on nontarget organisms. Here, we describe the successful use of the promoter and untranslated regions from the nanos (nos) orthologous gene of the yellow fever mosquito, Aedes aegypti, to control sex- and tissue-specific expression of exogenously derived mariner MosI transposase-encoding DNA. Transgenic mosquitoes expressed transposase mRNA in abundance near or equal to the endogenous nos transcript and exclusively in the female germ cells. In addition, MosI mRNA was deposited in developing oocytes and localized and maintained at the posterior pole during early embryonic development. Importantly, four of five transgenic lines examined were capable of mobilizing a second MosI transgene into the mosquito genome, indicating that functional transposase was being produced. Thus, the nos control sequences show promise as part of a TE-based gene drive system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

