Characterization of the physiological turnover of native and inactivated cytochromes P450 3A in cultured rat hepatocytes: a role for the cytosolic AAA ATPase p97?
- PMID: 17550236
- PMCID: PMC2536616
- DOI: 10.1021/bi700340n
Characterization of the physiological turnover of native and inactivated cytochromes P450 3A in cultured rat hepatocytes: a role for the cytosolic AAA ATPase p97?
Abstract
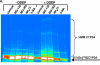
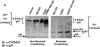
Mammalian hepatic cytochromes P450 (P450s) are endoplasmic reticulum (ER)-anchored hemoproteins engaged in the metabolism of numerous xeno- and endobiotics. P450s exhibit widely ranging half-lives, utilizing both autophagic-lysosomal (ALD) and ubiquitin-dependent 26S proteasomal (UPD) degradation pathways. Although suicidally inactivated hepatic CYPs 3A and "native" CYP3A4 in Saccharomyces cerevisiae are degraded via UPD, the turnover of native hepatic CYPs 3A in their physiological milieu has not been elucidated. Herein, we characterize the degradation of native, dexamethasone-inducible CYPs 3A in cultured primary rat hepatocytes, using proteasomal (MG-132 and MG-262) and ALD [NH4Cl and 3-methyladenine (3-MA)] inhibitors to examine their specific degradation route. Pulse-chase with immunoprecipitation analyses revealed a basal 52% 35S-CYP3A loss over 6 h, which was stabilized by both proteasomal inhibitors. By contrast, no corresponding CYP3A stabilization was detected with either ALD inhibitor NH4Cl or 3-MA. Furthermore, MG-262-induced CYP3A stabilization was associated with its polyubiquitylation, thereby verifying that native CYPs 3A were also degraded via UPD. To identify the specific participants in this process, cellular proteins were cross-linked in situ with paraformaldehyde (PFA) in cultured hepatocytes. Immunoblotting analyses of CYP3A immunoprecipitates after PFA-cross-linking revealed the presence of p97, a cytosolic AAA ATPase instrumental in the extraction and delivery of ubiquitylated ER proteins for proteasomal degradation. Such native CYP3A-p97 interactions were greatly magnified after CYP3A suicidal inactivation (which accelerates UPD), and/or proteasomal inhibition, and were confirmed by proteomic and confocal immunofluorescence microscopic analyses. These findings clearly reveal that native CYPs 3A undergo UPD and implicate a role for p97 in this process.
Figures












References
-
- Correia MA. Hepatic cytochrome P450 degradation: mechanistic diversity of the cellular sanitation brigade. Drug Metab Rev. 2003;35:107–143. and references therein. - PubMed
-
- Ohsumi Y. Molecular dissection of autophagy in the yeast Saccharomyces cerevisiae. In: Mayer RJ, Ciechanover A, Rechsteiner M, editors. Protein degradation. Vol. 2. The Ubiquitin-Proteasome system; 2006. 2006. pp. 31–50.
-
- Romisch K. Endoplasmic reticulum-associated degradation. Annu Rev Cell Dev Biol. 2005;21:435–456. - PubMed
-
- Schafer A, Wolf DH. Endoplasmic reticulum-associated protein quality control and degradation: genome-wide screen for ERAD components. Methods Mol Biol. 2005;301:289–292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

