Hepatitis C virus proteins
- PMID: 17552023
- PMCID: PMC4146758
- DOI: 10.3748/wjg.v13.i17.2406
Hepatitis C virus proteins
Abstract
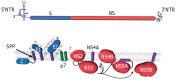
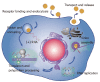
Hepatitis C virus (HCV) encodes a single polyprotein, which is processed by cellular and viral proteases to generate 10 polypeptides. The HCV genome also contains an overlapping +1 reading frame that may lead to the synthesis of an additional protein. Until recently, studies of HCV have been hampered by the lack of a productive cell culture system. Since the identification of HCV genome approximately 17 years ago, structural, biochemical and biological information on HCV proteins has mainly been obtained with proteins produced by heterologous expression systems. In addition, some functional studies have also been confirmed with replicon systems or with retroviral particles pseudotyped with HCV envelope glycoproteins. The data that have accumulated on HCV proteins begin to provide a framework for understanding the molecular mechanisms involved in the major steps of HCV life cycle. Moreover, the knowledge accumulated on HCV proteins is also leading to the development of antiviral drugs among which some are showing promising results in early-phase clinical trials. This review summarizes the current knowledge on the functions and biochemical features of HCV proteins.
Figures
References
-
- Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39:5–19. - PubMed
-
- Reed KE, Rice CM. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 2000;242:55–84. - PubMed
-
- Branch AD, Stump DD, Gutierrez JA, Eng F, Walewski JL. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin Liver Dis. 2005;25:105–117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

