Bidirectional, iterative approach to the structural delineation of the functional "chemoprint" in GPR40 for agonist recognition
- PMID: 17552505
- PMCID: PMC3592210
- DOI: 10.1021/jm0614782
Bidirectional, iterative approach to the structural delineation of the functional "chemoprint" in GPR40 for agonist recognition
Abstract
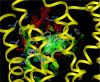
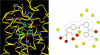
GPR40, free fatty acid receptor 1 (FFAR1), is a member of the GPCR superfamily and a possible target for the treatment of type 2 diabetes. In this work, we conducted a bidirectional iterative investigation, including computational modeling and site-directed mutagenesis, aimed at delineating amino acid residues forming the functional "chemoprint" of GPR40 for agonist recognition. The computational and experimental studies revolved around the recognition of the potent synthetic agonist GW9508. Our experimentally supported model suggested that H137(4.56), R183(5.39), N244(6.55), and R258(7.35) are directly involved in interactions with the ligand. We have proposed a polarized NH-pi interaction between H137(4.56) and GW9508 as one of the contributing forces leading to the high potency of GW9508. The modeling approach presented in this work provides a general strategy for the exploration of receptor-ligand interactions in G-protein coupled receptors beginning prior to acquisition of experimental data.
Figures



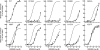


Similar articles
-
Identification of residues important for agonist recognition and activation in GPR40.J Biol Chem. 2007 Oct 5;282(40):29248-55. doi: 10.1074/jbc.M705077200. Epub 2007 Aug 15. J Biol Chem. 2007. PMID: 17699519
-
Cloning, identification and functional characterization of bovine free fatty acid receptor-1 (FFAR1/GPR40) in neutrophils.PLoS One. 2015 Mar 19;10(3):e0119715. doi: 10.1371/journal.pone.0119715. eCollection 2015. PLoS One. 2015. PMID: 25790461 Free PMC article.
-
Insight into analysis of interactions of GW9508 to wild-type and H86F and H137F GPR40: a combined QM/MM study and pharmacophore modeling.J Mol Graph Model. 2011 Apr;29(6):818-25. doi: 10.1016/j.jmgm.2011.01.006. Epub 2011 Feb 1. J Mol Graph Model. 2011. PMID: 21334233
-
Recent Updates on Free Fatty Acid Receptor 1 (GPR-40) Agonists for the Treatment of Type 2 Diabetes Mellitus.Mini Rev Med Chem. 2021;21(4):426-470. doi: 10.2174/1389557520666201023141326. Mini Rev Med Chem. 2021. PMID: 33100202 Review.
-
Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function.Pharmacol Ther. 2004 Jul;103(1):21-80. doi: 10.1016/j.pharmthera.2004.05.002. Pharmacol Ther. 2004. PMID: 15251227 Review.
Cited by
-
Molecular mechanism of fatty acid activation of FFAR1.Proc Natl Acad Sci U S A. 2023 May 30;120(22):e2219569120. doi: 10.1073/pnas.2219569120. Epub 2023 May 22. Proc Natl Acad Sci U S A. 2023. PMID: 37216523 Free PMC article.
-
Seven transmembrane-spanning receptors for free fatty acids as therapeutic targets for diabetes mellitus: pharmacological, phylogenetic, and drug discovery aspects.J Biol Chem. 2008 Jun 13;283(24):16269-73. doi: 10.1074/jbc.R800014200. Epub 2008 Apr 2. J Biol Chem. 2008. PMID: 18385136 Free PMC article. Review. No abstract available.
-
A novel antidiabetic drug, fasiglifam/TAK-875, acts as an ago-allosteric modulator of FFAR1.PLoS One. 2013 Oct 10;8(10):e76280. doi: 10.1371/journal.pone.0076280. eCollection 2013. PLoS One. 2013. PMID: 24130766 Free PMC article.
-
Novel selective ligands for free fatty acid receptors GPR120 and GPR40.Naunyn Schmiedebergs Arch Pharmacol. 2009 Sep;380(3):247-55. doi: 10.1007/s00210-009-0425-9. Epub 2009 May 27. Naunyn Schmiedebergs Arch Pharmacol. 2009. PMID: 19471906
-
Quantum mechanics implementation in drug-design workflows: does it really help?Drug Des Devel Ther. 2017 Aug 31;11:2551-2564. doi: 10.2147/DDDT.S126344. eCollection 2017. Drug Des Devel Ther. 2017. PMID: 28919707 Free PMC article. Review.
References
-
- Fanelli F, De Benedetti PG. Computational modeling approaches to structure-function analysis of G protein-coupled receptors. Chem. Rev. 2005;105:3297–3351. - PubMed
-
- Kristiansen K. Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol. Ther. 2004;103:21–80. - PubMed
-
- Lefkowitz RJ. Historical review: a brief history and personal retrospective of seven-transmembrane receptors. Trends Pharmacol. Sci. 2004;25:413–422. - PubMed
-
- Moro S, Spalluto G, Jacobson KA. Techniques: Recent developments in computer-aided engineering of GPCR ligands using the human adenosine A3 receptor as an example. Trends Pharmacol. Sci. 2005;26:44–51. - PubMed
-
- Bock JR, Gough DA. Virtual screen for ligands of orphan G protein-coupled receptors. J. Chem. Inf. Model. 2005;45:1402–1414. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases
Research Materials
Miscellaneous

