Robust interlaboratory reproducibility of a gene expression signature measurement consistent with the needs of a new generation of diagnostic tools
- PMID: 17553173
- PMCID: PMC1904205
- DOI: 10.1186/1471-2164-8-148
Robust interlaboratory reproducibility of a gene expression signature measurement consistent with the needs of a new generation of diagnostic tools
Abstract
Background: The increasing use of DNA microarrays in biomedical research, toxicogenomics, pharmaceutical development, and diagnostics has focused attention on the reproducibility and reliability of microarray measurements. While the reproducibility of microarray gene expression measurements has been the subject of several recent reports, there is still a need for systematic investigation into what factors most contribute to variability of measured expression levels observed among different laboratories and different experimenters.
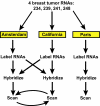
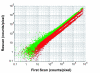
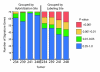
Results: We report the results of an interlaboratory comparison of gene expression array measurements on the same microarray platform, in which the RNA amplification and labeling, hybridization and wash, and slide scanning were each individually varied. Identical input RNA was used for all experiments. While some sources of variation have measurable influence on individual microarray signals, they showed very low influence on sample-to-reference ratios based on averaged triplicate measurements in the two-color experiments. RNA labeling was the largest contributor to interlaboratory variation.
Conclusion: Despite this variation, measurement of one particular breast cancer gene expression signature in three different laboratories was found to be highly robust, showing a high intralaboratory and interlaboratory reproducibility when using strictly controlled standard operating procedures.
Figures




References
-
- van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SL. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. - DOI - PubMed
-
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AAM, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. New Eng Jour Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. - DOI - PubMed
-
- Nutt CL, Mani DR, Betensky RA, Tamayo P, Cairncross JG, Ladd C, Pohl U, Hartmann C, McLaughlin ME, Batchelor TT, Black PM, von Deimling A, Pomeroy SL, Golub TR, Louis DN. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

