GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers
- PMID: 17553421
- PMCID: PMC2587162
- DOI: 10.1016/j.neuron.2007.05.022
GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers
Abstract
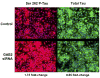
The apolipoprotein E (APOE) epsilon4 allele is the best established genetic risk factor for late-onset Alzheimer's disease (LOAD). We conducted genome-wide surveys of 502,627 single-nucleotide polymorphisms (SNPs) to characterize and confirm other LOAD susceptibility genes. In epsilon4 carriers from neuropathologically verified discovery, neuropathologically verified replication, and clinically characterized replication cohorts of 1411 cases and controls, LOAD was associated with six SNPs from the GRB-associated binding protein 2 (GAB2) gene and a common haplotype encompassing the entire GAB2 gene. SNP rs2373115 (p = 9 x 10(-11)) was associated with an odds ratio of 4.06 (confidence interval 2.81-14.69), which interacts with APOE epsilon4 to further modify risk. GAB2 was overexpressed in pathologically vulnerable neurons; the Gab2 protein was detected in neurons, tangle-bearing neurons, and dystrophic neuritis; and interference with GAB2 gene expression increased tau phosphorylation. Our findings suggest that GAB2 modifies LOAD risk in APOE epsilon4 carriers and influences Alzheimer's neuropathology.
Figures


References
-
- Ahmadian A, Gharizadeh B, Gustafsson AC, Sterky F, Nyren P, Uhlen M, Lundeberg J. Single-nucleotide polymorphism analysis by pyrosequencing. Anal Biochem. 2000;280:103–110. - PubMed
-
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. - PubMed
-
- Bobinski M, de Leon MJ, Wegiel J, Desanti S, Convit A, Saint Louis LA, Rusinek H, Wisniewski HM. The histological validation of post mortem magnetic resonance imaging-determined hippocampal volume in Alzheimer’s disease. Neuroscience. 2000;95:721–725. - PubMed
-
- Braak H, Braak E. Neuropathological staging of Alzheimer’s-related changes. Acta Neuropathol (Berl) 1991;82:239–259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 AG005128/AG/NIA NIH HHS/United States
- R01 MH60451/MH/NIMH NIH HHS/United States
- P30 AG013846/AG/NIA NIH HHS/United States
- P30 AG10161/AG/NIA NIH HHS/United States
- P50 AG25711/AG/NIA NIH HHS/United States
- P30 AG19610/AG/NIA NIH HHS/United States
- P50 AG08702/AG/NIA NIH HHS/United States
- P30-AG12300/AG/NIA NIH HHS/United States
- P30 AG13846/AG/NIA NIH HHS/United States
- R01 AG023193/AG/NIA NIH HHS/United States
- R21 AG029576/AG/NIA NIH HHS/United States
- P50 AG16574/AG/NIA NIH HHS/United States
- P50 MH060451/MH/NIMH NIH HHS/United States
- P50 AG17824/AG/NIA NIH HHS/United States
- P50 AG08671/AG/NIA NIH HHS/United States
- P50 AG 05681/AG/NIA NIH HHS/United States
- P50 AG16570/AG/NIA NIH HHS/United States
- P50 AG008671/AG/NIA NIH HHS/United States
- P50 AG05136/AG/NIA NIH HHS/United States
- P50 AG005142/AG/NIA NIH HHS/United States
- U24NS051872/NS/NINDS NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- P50 AG005146/AG/NIA NIH HHS/United States
- U01HL084744/HL/NHLBI NIH HHS/United States
- G0701075/MRC_/Medical Research Council/United Kingdom
- P50 AG05146/AG/NIA NIH HHS/United States
- P50 AG05142/AG/NIA NIH HHS/United States
- U24 NS051872/NS/NINDS NIH HHS/United States
- K01 AG024079/AG/NIA NIH HHS/United States
- P50 AG008702/AG/NIA NIH HHS/United States
- P30 AG017824/AG/NIA NIH HHS/United States
- U01 AG016976/AG/NIA NIH HHS/United States
- P50 NS039764/NS/NINDS NIH HHS/United States
- P50 AG005681/AG/NIA NIH HHS/United States
- P50 AG05134/AG/NIA NIH HHS/United States
- P50 AG005136/AG/NIA NIH HHS/United States
- P30 AG012300/AG/NIA NIH HHS/United States
- R01 AG23193/AG/NIA NIH HHS/United States
- K01AG024079/AG/NIA NIH HHS/United States
- P50 AG016570/AG/NIA NIH HHS/United States
- P50 AG005134/AG/NIA NIH HHS/United States
- R01 NS39764/NS/NINDS NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- P50 AG05144/AG/NIA NIH HHS/United States
- AG025688/AG/NIA NIH HHS/United States
- P50 AG025688/AG/NIA NIH HHS/United States
- P50 AG005144/AG/NIA NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- P30 AG01542/AG/NIA NIH HHS/United States
- P50 AG05128/AG/NIA NIH HHS/United States
- P50 AG025711/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

