Hydrophobic residues that form putative fusion loops of Epstein-Barr virus glycoprotein B are critical for fusion activity
- PMID: 17553877
- PMCID: PMC1951416
- DOI: 10.1128/JVI.00758-07
Hydrophobic residues that form putative fusion loops of Epstein-Barr virus glycoprotein B are critical for fusion activity
Abstract
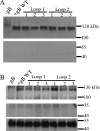
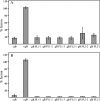
To test the importance of the hydrophobic residues within the putative Epstein-Barr virus (EBV) glycoprotein B (gB) fusion loops in membrane fusion, WY(112-113) and WLIW(193-196) were mutated into alanine, glutamic acid, or the analogous residues from herpes simplex virus type 1 (HSV-1) gB (HR and RVEA). All gB variants exhibited cell surface expression, demonstrating that the substitutions did not perturb gB trafficking. None of six gB variants was, however, capable of mediating fusion with either epithelial or B cells. These data demonstrate that the bulky and hydrophobic EBV loop residues, which differ from the more hydrophilic HSV-1 residues and appear more compatible with membrane insertion, are essential for EBV gB-dependent fusion.
Figures


References
-
- Burnette, W. N. 1981. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112:195-203. - PubMed
-
- Daniels, G. M., and S. G. Amara. 1998. Selective labeling of neurotransmitter transporters at the cell surface. Methods Enzymol. 296:307-318. - PubMed
-
- DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
-
- Dutch, R. E., T. S. Jardetzky, and R. A. Lamb. 2000. Virus membrane fusion proteins: biological machines that undergo a metamorphosis. Biosci. Rep. 20:597-612. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

