Recombination confounds the early evolutionary history of human immunodeficiency virus type 1: subtype G is a circulating recombinant form
- PMID: 17553886
- PMCID: PMC1951349
- DOI: 10.1128/JVI.00463-07
Recombination confounds the early evolutionary history of human immunodeficiency virus type 1: subtype G is a circulating recombinant form
Abstract
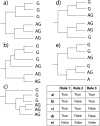
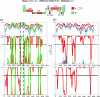
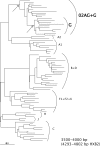
Human immunodeficiency virus type 1 (HIV-1) is classified in nine subtypes (A to D, F, G, H, J, and K), a number of subsubtypes, and several circulating recombinant forms (CRFs). Due to the high level of genetic diversity within HIV-1 and to its worldwide distribution, this classification system is widely used in fields as diverse as vaccine development, evolution, epidemiology, viral fitness, and drug resistance. Here, we demonstrate how the high recombination rates of HIV-1 may confound the study of its evolutionary history and classification. Our data show that subtype G, currently classified as a pure subtype, has in fact a recombinant history, having evolved following recombination between subtypes A and J and a putative subtype G parent. In addition, we find no evidence for recombination within one of the lineages currently classified as a CRF, CRF02_AG. Our analysis indicates that CRF02_AG was the parent of the recombinant subtype G, rather than the two having the opposite evolutionary relationship, as is currently proposed. Our results imply that the current classification of HIV-1 subtypes and CRFs is an artifact of sampling history, rather than reflecting the evolutionary history of the virus. We suggest a reanalysis of all pure subtypes and CRFs in order to better understand how high rates of recombination have influenced HIV-1 evolutionary history.
Figures


Similar articles
-
Identification of a novel circulating recombinant form (CRF) 36_cpx in Cameroon that combines two CRFs (01_AE and 02_AG) with ancestral lineages of subtypes A and G.AIDS Res Hum Retroviruses. 2007 Aug;23(8):1008-19. doi: 10.1089/aid.2006.0289. AIDS Res Hum Retroviruses. 2007. PMID: 17725418
-
Identification of new CRF43_02G and CRF25_cpx in Saudi Arabia based on full genome sequence analysis of six HIV type 1 isolates.AIDS Res Hum Retroviruses. 2008 Oct;24(10):1327-35. doi: 10.1089/aid.2008.0101. AIDS Res Hum Retroviruses. 2008. PMID: 18844465
-
Rare HIV-1 Subtype J Genomes and a New H/U/CRF02_AG Recombinant Genome Suggests an Ancient Origin of HIV-1 in Angola.AIDS Res Hum Retroviruses. 2016 Aug;32(8):822-8. doi: 10.1089/AID.2016.0084. Epub 2016 Jun 1. AIDS Res Hum Retroviruses. 2016. PMID: 27098898 Free PMC article.
-
[Genetic diversity of HIV infection worldwide and its consequences].Med Trop (Mars). 1999;59(4 Pt 2):449-55. Med Trop (Mars). 1999. PMID: 10901846 Review. French.
-
Genetic diversity of human immunodeficiency virus-1 in Nigeria: 2002-2017 - systematic review and meta-analysis.Niger Postgrad Med J. 2020 Jan-Mar;27(1):8-12. doi: 10.4103/npmj.npmj_64_19. Niger Postgrad Med J. 2020. PMID: 32003356
Cited by
-
Characterization of HIV-1 gag and nef in Cameroon: further evidence of extreme diversity at the origin of the HIV-1 group M epidemic.Virol J. 2013 Jan 22;10:29. doi: 10.1186/1743-422X-10-29. Virol J. 2013. PMID: 23339631 Free PMC article.
-
Revisiting the recombinant history of HIV-1 group M with dynamic network community detection.Proc Natl Acad Sci U S A. 2022 May 10;119(19):e2108815119. doi: 10.1073/pnas.2108815119. Epub 2022 May 2. Proc Natl Acad Sci U S A. 2022. PMID: 35500121 Free PMC article.
-
The substitution rate of HIV-1 subtypes: a genomic approach.Virus Evol. 2017 Oct 20;3(2):vex029. doi: 10.1093/ve/vex029. eCollection 2017 Jul. Virus Evol. 2017. PMID: 29942652 Free PMC article.
-
Primate immunodeficiency virus classification and nomenclature: Review.Infect Genet Evol. 2016 Dec;46:150-158. doi: 10.1016/j.meegid.2016.10.018. Epub 2016 Oct 24. Infect Genet Evol. 2016. PMID: 27789390 Free PMC article. Review.
-
Near full-length HIV type 1M genomic sequences from Cameroon : Evidence of early diverging under-sampled lineages in the country.Evol Med Public Health. 2015 Sep 9;2015(1):254-65. doi: 10.1093/emph/eov022. Evol Med Public Health. 2015. PMID: 26354000 Free PMC article.
References
-
- Carr, J. K., T. Laukkanen, M. O. Salminen, J. Albert, A. Alaeus, B. Kim, E. Sanders-Buell, D. L. Birx, and F. E. McCutchan. 1999. Characterization of subtype A HIV-1 from Africa by full genome sequencing. AIDS 13:1819-1826. - PubMed
-
- Carr, J. K., M. O. Salminen, J. Albert, E. Sanders-Buell, D. Gotte, D. L. Birx, and F. E. McCutchan. 1998. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology 247:22-31. - PubMed
-
- de Oliveira, T., K. Deforche, S. Cassol, M. Salminen, D. Paraskevis, C. Seebregts, J. Snoeck, E. J. van Rensburg, A. M. Wensing, D. A. van de Vijver, C. A. Boucher, R. Camacho, and A. M. Vandamme. 2005. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 21:3797-3800. - PubMed
-
- Fonjungo, P. N., E. N. Mpoudi, J. N. Torimiro, G. A. Alemnji, L. T. Eno, E. J. Lyonga, J. N. Nkengasong, R. B. Lal, M. Rayfield, M. L. Kalish, T. M. Folks, and D. Pieniazek. 2002. Human immunodeficiency virus type 1 group M protease in Cameroon: genetic diversity and protease inhibitor mutational features. J. Clin. Microbiol. 40:837-845. - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

