Riboswitching on RNA virus replication
- PMID: 17553958
- PMCID: PMC1965526
- DOI: 10.1073/pnas.0704178104
Riboswitching on RNA virus replication
Abstract
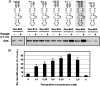
Positive-strand RNA viruses direct different virus-specific processes during their infection of host cells. Fundamental events such as viral RNA genome replication are controlled by viral regulatory RNA elements (REs). Here, we have investigated the possibility of specifically modulating the action of a viral RE using RNA aptamer technology. Through rational design, a tombusvirus RE, which has the structure of a perfect RNA stem loop in the plus-strand RNA genome, was replaced with a theophylline-binding RNA aptamer sequence, an imperfect stem loop. The aptamer-RE hybrid was designed so that, upon binding theophylline, it would become more stable and structurally mimic the functional RE (i.e., represent a ligand-inducible RE riboswitch). Initial experiments were conducted with a small noncoding virus genome-derived RNA replicon, and the results showed that replication was inducible, up to approximately 10-fold, in a theophylline-specific and dose-dependent manner. A similar level of theophylline-dependent induction was also observed when a full-length viral genome containing an RE riboswitch was tested. Analysis of this engineered viral genome revealed that this RE, located in the 5' untranslated region, specifically mediates efficient accumulation of plus-strands of the virus genome. Therefore, in addition to allowing for modulation of virus reproduction, the RE riboswitch system also provided insight into RE function. The ability to chemically induce a viral process via modulation of virus genome structure could be useful for basic and applied aspects of research.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
An RNA domain within the 5' untranslated region of the tomato bushy stunt virus genome modulates viral RNA replication.J Mol Biol. 2001 Jan 26;305(4):741-56. doi: 10.1006/jmbi.2000.4298. J Mol Biol. 2001. PMID: 11162089
-
The 5'-terminal region of the Aichi virus genome encodes cis-acting replication elements required for positive- and negative-strand RNA synthesis.J Virol. 2005 Jun;79(11):6918-31. doi: 10.1128/JVI.79.11.6918-6931.2005. J Virol. 2005. PMID: 15890931 Free PMC article.
-
Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication.J Virol. 2005 Apr;79(8):4859-69. doi: 10.1128/JVI.79.8.4859-4869.2005. J Virol. 2005. PMID: 15795271 Free PMC article.
-
Replicon-based vectors of positive strand RNA viruses.Curr Opin Mol Ther. 2000 Oct;2(5):555-69. Curr Opin Mol Ther. 2000. PMID: 11249758 Review.
-
Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication.Virology. 2006 Jan 5;344(1):211-20. doi: 10.1016/j.virol.2005.09.017. Virology. 2006. PMID: 16364751 Review.
Cited by
-
Rational design of artificial riboswitches based on ligand-dependent modulation of internal ribosome entry in wheat germ extract and their applications as label-free biosensors.RNA. 2011 Mar;17(3):478-88. doi: 10.1261/rna.2433111. Epub 2011 Jan 11. RNA. 2011. PMID: 21224378 Free PMC article.
-
A discontinuous RNA platform mediates RNA virus replication: building an integrated model for RNA-based regulation of viral processes.PLoS Pathog. 2009 Mar;5(3):e1000323. doi: 10.1371/journal.ppat.1000323. Epub 2009 Mar 6. PLoS Pathog. 2009. PMID: 19266082 Free PMC article.
-
Subgenomic messenger RNAs: mastering regulation of (+)-strand RNA virus life cycle.Virology. 2011 Apr 10;412(2):245-55. doi: 10.1016/j.virol.2011.02.007. Epub 2011 Mar 5. Virology. 2011. PMID: 21377709 Free PMC article. Review.
-
Intergenerational phenotypic mixing in viral evolution.Evolution. 2013 Jun;67(6):1815-22. doi: 10.1111/evo.12048. Epub 2013 Feb 4. Evolution. 2013. PMID: 23730772 Free PMC article.
-
Advances in RNA structure prediction from sequence: new tools for generating hypotheses about viral RNA structure-function relationships.J Virol. 2009 Jul;83(13):6326-34. doi: 10.1128/JVI.00251-09. Epub 2009 Apr 15. J Virol. 2009. PMID: 19369331 Free PMC article. Review. No abstract available.
References
-
- Jenison RD, Gill SC, Pardi A, Polisky B. Science. 1994;263:1425–1429. - PubMed
-
- Winkler WC, Breaker RR. ChemBioChem. 2003;4:1024–1032. - PubMed
-
- Isaacs FJ, Dwyer DJ, Collins JJ. Nat Biotechnol. 2006;24:545–554. - PubMed
-
- Werstuck G, Green MR. Science. 1998;282:296–298. - PubMed
-
- Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ. Nat Biotechnol. 2004;22:841–847. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

