Dissociation of halted T7 RNA polymerase elongation complexes proceeds via a forward-translocation mechanism
- PMID: 17553968
- PMCID: PMC1965517
- DOI: 10.1073/pnas.0606306104
Dissociation of halted T7 RNA polymerase elongation complexes proceeds via a forward-translocation mechanism
Abstract
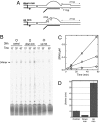
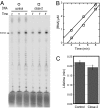
A recent model for the mechanism of intrinsic transcription termination involves dissociation of the RNA from forward-translocated (hypertranslocated) states of the complex [Yarnell WS, Roberts JW (1999) Science, 284:611-615]. The current study demonstrates that halted elongation complexes of T7 RNA polymerase in the absence of termination signals can also dissociate via a forward-translocation mechanism. Shortening of the downstream DNA or the introduction of a stretch of mismatched DNA immediately downstream of the halt site reduces a barrier to forward translocation and correspondingly reduces the lifetime of halted complexes. Conversely, introduction of a cross-link downstream of the halt site increases the same barrier and leads to an increase in complex lifetime. Introduction of a mismatch within the bubble reduces a driving force for forward translocation and correspondingly increases the lifetime of the complex, but only for mismatches at the upstream edge of the bubble, as predicted by the model. Mismatching only the two most upstream of the eight bases in the bubble provides a maximal increase in complex stability, suggesting that dissociation occurs primarily from early forward-translocated states. Finally, addition in trans of an oligonucleotide complementary to the nascent RNA just beyond the hybrid complements the loss of driving force derived from placement of a mismatch within the bubble, confirming the expected additivity of effects. Thus, forward translocation is likely a general mechanism for dissociation of elongation complexes, both in the presence and absence of intrinsic termination signals.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Characterization of halted T7 RNA polymerase elongation complexes reveals multiple factors that contribute to stability.J Mol Biol. 2000 Oct 6;302(5):1049-62. doi: 10.1006/jmbi.2000.4114. J Mol Biol. 2000. PMID: 11183774
-
Studies on the interaction of T7 RNA polymerase with a DNA template containing a site-specifically placed psoralen cross-link. II. Stability and some properties of elongation complexes.J Mol Biol. 1991 Oct 20;221(4):1111-25. J Mol Biol. 1991. PMID: 1942045
-
Fluorescence characterization of the transcription bubble in elongation complexes of T7 RNA polymerase.J Mol Biol. 2001 May 4;308(3):465-75. doi: 10.1006/jmbi.2001.4601. J Mol Biol. 2001. PMID: 11327781
-
Studies on the interaction of T7 RNA polymerase with a DNA template containing a site-specifically placed psoralen cross-link. I. Characterization of elongation complexes.J Mol Biol. 1991 Oct 20;221(4):1091-110. J Mol Biol. 1991. PMID: 1942044
-
Structure and function in promoter escape by T7 RNA polymerase.Prog Nucleic Acid Res Mol Biol. 2005;80:323-47. doi: 10.1016/S0079-6603(05)80008-X. Prog Nucleic Acid Res Mol Biol. 2005. PMID: 16164978 Review. No abstract available.
Cited by
-
Single-mode termination of phage transcriptions, disclosing bacterial adaptation for facilitated reinitiations.Nucleic Acids Res. 2024 Aug 27;52(15):9092-9102. doi: 10.1093/nar/gkae620. Nucleic Acids Res. 2024. PMID: 39011892 Free PMC article.
-
Archaeal intrinsic transcription termination in vivo.J Bacteriol. 2009 Nov;191(22):7102-8. doi: 10.1128/JB.00982-09. Epub 2009 Sep 11. J Bacteriol. 2009. PMID: 19749050 Free PMC article.
-
3' end additions by T7 RNA polymerase are RNA self-templated, distributive and diverse in character-RNA-Seq analyses.Nucleic Acids Res. 2018 Oct 12;46(18):9253-9263. doi: 10.1093/nar/gky796. Nucleic Acids Res. 2018. PMID: 30219859 Free PMC article.
-
RNA polymerase II flexibility during translocation from normal mode analysis.Proteins. 2010 Feb 1;78(2):434-46. doi: 10.1002/prot.22560. Proteins. 2010. PMID: 19714773 Free PMC article.
-
A Viral T7 RNA Polymerase Ratcheting Along DNA With Fidelity Control.Comput Struct Biotechnol J. 2019 May 9;17:638-644. doi: 10.1016/j.csbj.2019.05.001. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 31193497 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

