Sponge-associated microorganisms: evolution, ecology, and biotechnological potential
- PMID: 17554047
- PMCID: PMC1899876
- DOI: 10.1128/MMBR.00040-06
Sponge-associated microorganisms: evolution, ecology, and biotechnological potential
Abstract
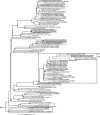
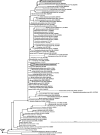
Marine sponges often contain diverse and abundant microbial communities, including bacteria, archaea, microalgae, and fungi. In some cases, these microbial associates comprise as much as 40% of the sponge volume and can contribute significantly to host metabolism (e.g., via photosynthesis or nitrogen fixation). We review in detail the diversity of microbes associated with sponges, including extensive 16S rRNA-based phylogenetic analyses which support the previously suggested existence of a sponge-specific microbiota. These analyses provide a suitable vantage point from which to consider the potential evolutionary and ecological ramifications of these widespread, sponge-specific microorganisms. Subsequently, we examine the ecology of sponge-microbe associations, including the establishment and maintenance of these sometimes intimate partnerships, the varied nature of the interactions (ranging from mutualism to host-pathogen relationships), and the broad-scale patterns of symbiont distribution. The ecological and evolutionary importance of sponge-microbe associations is mirrored by their enormous biotechnological potential: marine sponges are among the animal kingdom's most prolific producers of bioactive metabolites, and in at least some cases, the compounds are of microbial rather than sponge origin. We review the status of this important field, outlining the various approaches (e.g., cultivation, cell separation, and metagenomics) which have been employed to access the chemical wealth of sponge-microbe associations.
Figures











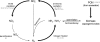


References
-
- Adamczeski, M., A. R. Reed, and P. Crews. 1995. New and known diketopiperazines from the Caribbean sponge, Calyx cf. podatypa. J. Nat. Prod. 58:201-208. - PubMed
-
- Adamczyk, J., M. Hesselsoe, N. Iversen, M. Horn, A. Lehner, P. H. Nielsen, M. Schloter, P. Roslev, and M. Wagner. 2003. The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Appl. Environ. Microbiol. 69:6875-6887. - PMC - PubMed
-
- Aicher, T. D., K. R. Buszek, F. G. Fang, C. J. Forsyth, S. H. Jung, Y. Kishi, M. C. Matelich, P. M. Scola, D. M. Spero, and S. K. Yoon. 1992. Total synthesis of halichondrin B and norhalichondrin B. J. Am. Chem. Soc. 114:3162-3164.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

