Listeria monocytogenes surface proteins: from genome predictions to function
- PMID: 17554049
- PMCID: PMC1899877
- DOI: 10.1128/MMBR.00039-06
Listeria monocytogenes surface proteins: from genome predictions to function
Abstract
The genome of the human food-borne pathogen Listeria monocytogenes is predicted to encode a high number of surface proteins. This abundance likely reflects the ability of this bacterium to survive in diverse environments, including soil, food, and the human host. This review focuses on the various mechanisms by which listerial proteins are attached at the bacterial surface and their many functions, including peptidoglycan metabolism, protein processing, adhesion to host cells, and invasion of host tissues. Extensive in silico analysis of the domains or motifs present in these mosaic proteins reveals that diverse structural features allow the surface proteome to interact with diverse bacterial or host components. This diversity offers new clues about the molecular bases of Listeria pathogenesis.
Figures
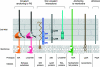



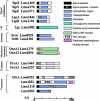
Similar articles
-
Genetic diversity of Listeria monocytogenes recovered from infected persons and pork, seafood and dairy products on retail sale in France during 2000 and 2001.Int J Food Microbiol. 2007 Mar 10;114(2):187-94. doi: 10.1016/j.ijfoodmicro.2006.09.011. Epub 2006 Dec 26. Int J Food Microbiol. 2007. PMID: 17188773
-
Comparative genomics of Listeria species.Science. 2001 Oct 26;294(5543):849-52. doi: 10.1126/science.1063447. Science. 2001. PMID: 11679669
-
The protein secretion systems in Listeria: inside out bacterial virulence.FEMS Microbiol Rev. 2006 Sep;30(5):774-805. doi: 10.1111/j.1574-6976.2006.00035.x. FEMS Microbiol Rev. 2006. PMID: 16911044 Review.
-
Listeria monocytogenes: a multifaceted model.Nat Rev Microbiol. 2006 Jun;4(6):423-34. doi: 10.1038/nrmicro1413. Nat Rev Microbiol. 2006. PMID: 16710323 Review.
-
Classes and functions of Listeria monocytogenes surface proteins.Pol J Microbiol. 2004;53(2):75-88. Pol J Microbiol. 2004. PMID: 15478352 Review.
Cited by
-
The Mucus Binding Factor Is Not Necessary for Lacticaseibacillus rhamnosus CRL1505 to Exert Its Immunomodulatory Activities in Local and Distal Mucosal Sites.Int J Mol Sci. 2022 Nov 18;23(22):14357. doi: 10.3390/ijms232214357. Int J Mol Sci. 2022. PMID: 36430834 Free PMC article.
-
Suppressor Mutations Linking gpsB with the First Committed Step of Peptidoglycan Biosynthesis in Listeria monocytogenes.J Bacteriol. 2016 Dec 13;199(1):e00393-16. doi: 10.1128/JB.00393-16. Print 2017 Jan 1. J Bacteriol. 2016. PMID: 27795316 Free PMC article.
-
Listeria monocytogenes-How This Pathogen Uses Its Virulence Mechanisms to Infect the Hosts.Pathogens. 2022 Dec 7;11(12):1491. doi: 10.3390/pathogens11121491. Pathogens. 2022. PMID: 36558825 Free PMC article. Review.
-
Genetic Separation of Listeria monocytogenes Causing Central Nervous System Infections in Animals.Front Cell Infect Microbiol. 2018 Feb 5;8:20. doi: 10.3389/fcimb.2018.00020. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29459888 Free PMC article.
-
Actin polymerization drives septation of Listeria monocytogenes namA hydrolase mutants, demonstrating host correction of a bacterial defect.Infect Immun. 2011 Apr;79(4):1458-70. doi: 10.1128/IAI.01140-10. Epub 2011 Jan 24. Infect Immun. 2011. PMID: 21263016 Free PMC article.
References
-
- Abachin, E., C. Poyart, E. Pellegrini, E. Milohanic, F. Fiedler, P. Berche, and P. Trieu-Cuot. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 43:1-14. - PubMed
-
- Alvarez-Dominguez, C., J. A. Vazquez-Boland, E. Carrasco-Marin, P. Lopez-Mato, and F. Leyva-Cobian. 1997. Host cell heparan sulfate proteoglycans mediate attachment and entry of Listeria monocytogenes, and the listerial surface protein ActA is involved in heparan sulfate receptor recognition. Infect. Immun. 65:78-88. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

