Viral proteomics
- PMID: 17554050
- PMCID: PMC1899879
- DOI: 10.1128/MMBR.00042-06
Viral proteomics
Erratum in
- Microbiol Mol Biol Rev. 2007 Sep;71(3):549
Abstract
Viruses have long been studied not only for their pathology and associated disease but also as model systems for molecular processes and as tools for identifying important cellular regulatory proteins and pathways. Recent advances in mass spectrometry methods coupled with the development of proteomic approaches have greatly facilitated the detection of virion components, protein interactions in infected cells, and virally induced changes in the cellular proteome, resulting in a more comprehensive understanding of viral infection. In addition, a rapidly increasing number of high-resolution structures for viral proteins have provided valuable information on the mechanism of action of these proteins as well as aided in the design and understanding of specific inhibitors that could be used in antiviral therapies. In this paper, we discuss proteomic studies conducted on all eukaryotic viruses and bacteriophages, covering virion composition, viral protein structures, virus-virus and virus-host protein interactions, and changes in the cellular proteome upon viral infection.
Figures
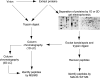
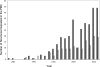


References
-
- Alfonso, P., J. Rivera, B. Hernaez, C. Alonso, and J. M. Escribano. 2004. Identification of cellular proteins modified in response to African swine fever virus infection by proteomics. Proteomics 4:2037-2046. - PubMed
-
- Anderson, W. F., D. H. Ohlendorf, Y. Takeda, and B. W. Matthews. 1981. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature 290:754-758. - PubMed
-
- Baas, T., C. R. Baskin, D. L. Diamond, A. Garcia-Sastre, H. Bielefeldt-Ohmann, T. M. Tumpey, M. J. Thomas, V. S. Carter, T. H. Teal, N. Van Hoeven, S. Proll, J. M. Jacobs, Z. R. Caldwell, M. A. Gritsenko, R. R. Hukkanen, D. G. Camp II, R. D. Smith, and M. G. Katze. 2006. Integrated molecular signature of disease: analysis of influenza virus-infected macaques through functional genomics and proteomics. J. Virol. 80:10813-10828. - PMC - PubMed
-
- Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. S. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

