Identification of sequence motifs significantly associated with antisense activity
- PMID: 17555590
- PMCID: PMC1919396
- DOI: 10.1186/1471-2105-8-184
Identification of sequence motifs significantly associated with antisense activity
Abstract
Background: Predicting the suppression activity of antisense oligonucleotide sequences is the main goal of the rational design of nucleic acids. To create an effective predictive model, it is important to know what properties of an oligonucleotide sequence associate significantly with antisense activity. Also, for the model to be efficient we must know what properties do not associate significantly and can be omitted from the model. This paper will discuss the results of a randomization procedure to find motifs that associate significantly with either high or low antisense suppression activity, analysis of their properties, as well as the results of support vector machine modelling using these significant motifs as features.
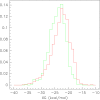
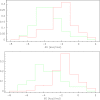
Results: We discovered 155 motifs that associate significantly with high antisense suppression activity and 202 motifs that associate significantly with low suppression activity. The motifs range in length from 2 to 5 bases, contain several motifs that have been previously discovered as associating highly with antisense activity, and have thermodynamic properties consistent with previous work associating thermodynamic properties of sequences with their antisense activity. Statistical analysis revealed no correlation between a motif's position within an antisense sequence and that sequences antisense activity. Also, many significant motifs existed as subwords of other significant motifs. Support vector regression experiments indicated that the feature set of significant motifs increased correlation compared to all possible motifs as well as several subsets of the significant motifs.
Conclusion: The thermodynamic properties of the significantly associated motifs support existing data correlating the thermodynamic properties of the antisense oligonucleotide with antisense efficiency, reinforcing our hypothesis that antisense suppression is strongly associated with probe/target thermodynamics, as there are no enzymatic mediators to speed the process along like the RNA Induced Silencing Complex (RISC) in RNAi. The independence of motif position and antisense activity also allows us to bypass consideration of this feature in the modelling process, promoting model efficiency and reducing the chance of overfitting when predicting antisense activity. The increase in SVR correlation with significant features compared to nearest-neighbour features indicates that thermodynamics alone is likely not the only factor in determining antisense efficiency.
Figures

Similar articles
-
Computational models with thermodynamic and composition features improve siRNA design.BMC Bioinformatics. 2006 Feb 12;7:65. doi: 10.1186/1471-2105-7-65. BMC Bioinformatics. 2006. PMID: 16472402 Free PMC article.
-
A Gibbs sampler for identification of symmetrically structured, spaced DNA motifs with improved estimation of the signal length.Bioinformatics. 2005 May 15;21(10):2240-5. doi: 10.1093/bioinformatics/bti336. Epub 2005 Feb 22. Bioinformatics. 2005. PMID: 15728117
-
Generalized hierarchical markov models for the discovery of length-constrained sequence features from genome tiling arrays.Biometrics. 2007 Sep;63(3):797-805. doi: 10.1111/j.1541-0420.2007.00760.x. Biometrics. 2007. PMID: 17825011
-
Antisense transgenics in animals.Methods. 1999 Jul;18(3):304-10. doi: 10.1006/meth.1999.0788. Methods. 1999. PMID: 10454989 Review.
-
[Antisense strategies come of age].Tanpakushitsu Kakusan Koso. 1993 Mar;38(4):753-65. Tanpakushitsu Kakusan Koso. 1993. PMID: 8475331 Review. Japanese. No abstract available.
Cited by
-
Antisense DNA parameters derived from next-nearest-neighbor analysis of experimental data.BMC Bioinformatics. 2010 May 14;11:252. doi: 10.1186/1471-2105-11-252. BMC Bioinformatics. 2010. PMID: 20470414 Free PMC article.
-
Gene silencing by synthetic U1 adaptors.Nat Biotechnol. 2009 Mar;27(3):257-63. doi: 10.1038/nbt.1525. Epub 2009 Feb 15. Nat Biotechnol. 2009. PMID: 19219028
-
Increased in vivo inhibition of gene expression by combining RNA interference and U1 inhibition.Nucleic Acids Res. 2012 Jan;40(1):e8. doi: 10.1093/nar/gkr956. Epub 2011 Nov 15. Nucleic Acids Res. 2012. PMID: 22086952 Free PMC article.
-
Knockdown of LdMC1 and Hsp70 by antisense oligonucleotides causes cell-cycle defects and programmed cell death in Leishmania donovani.Mol Cell Biochem. 2012 Jan;359(1-2):135-49. doi: 10.1007/s11010-011-1007-y. Epub 2011 Jul 31. Mol Cell Biochem. 2012. PMID: 21805355
-
IDT SciTools: a suite for analysis and design of nucleic acid oligomers.Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W163-9. doi: 10.1093/nar/gkn198. Epub 2008 Apr 25. Nucleic Acids Res. 2008. PMID: 18440976 Free PMC article.
References
-
- Crooke ST. Molecular mechanisms of action of antisense drugs. Biochim Biophys Acta. 1999;1489:31–44. - PubMed
-
- Crooke ST. Molecular mechanisms of antisense drugs: RNase H. Antisense Nucleic Acid Drug Dev. 1998;8:133–134. - PubMed
-
- Jayaraman A, Walton SP, Yarmush ML, Roth CM. Rational selection and quantitative evaluation of antisense oligonucleotides. Biochim Biophys Acta. 2001;1520:105–114. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

