The rate of TRAP binding to RNA is crucial for transcription attenuation control of the B. subtilis trp operon
- PMID: 17555767
- PMCID: PMC2034321
- DOI: 10.1016/j.jmb.2007.05.027
The rate of TRAP binding to RNA is crucial for transcription attenuation control of the B. subtilis trp operon
Abstract
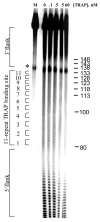
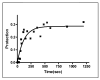
The trp RNA-binding attenuation protein (TRAP) regulates expression of the tryptophan biosynthetic and transport genes in Bacillus subtilis in response to changes in the levels of intracellular tryptophan. Transcription of the trpEDCFBA operon is controlled by an attenuation mechanism involving two overlapping RNA secondary structures in the 5' leader region of the trp transcript; TRAP binding promotes formation of a transcription terminator structure that halts transcription prior to the structural genes. TRAP consists of 11 identical subunits and is activated to bind RNA by binding up to 11 molecules of L-tryptophan. The TRAP binding site in the leader region of the trp operon mRNA consists of 11 (G/U)AG repeats. We examined the importance of the rate of TRAP binding to RNA for the transcription attenuation mechanism. We compared the properties of two types of TRAP 11-mers: homo-11-mers composed of 11 wild-type subunits, and hetero-11-mers with only one wild-type subunit and ten mutant subunits defective in binding either RNA or tryptophan. The hetero-11-mers bound RNA with only slightly diminished equilibrium binding affinity but with slower on-rates as compared to WT TRAP. The hetero-11-mers showed significantly decreased ability to induce transcription termination in the trp leader region when examined using an in vitro attenuation system. Together these results indicate that the rate of TRAP binding to RNA is a crucial factor in TRAP's ability to control attenuation.
Figures
















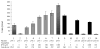
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

