Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila
- PMID: 17555964
- PMCID: PMC1963421
- DOI: 10.1016/j.cub.2007.05.039
Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila
Abstract
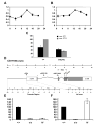
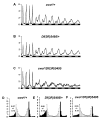
Gene transcription is a central timekeeping process in animal clocks. In Drosophila, the basic helix-loop helix (bHLH)-PAS transcription-factor heterodimer, CLOCK/CYCLE (CLK/CYC), transcriptionally activates the clock components period (per), timeless (tim), Par domain protein 1 (Pdp1), and vrille (vri), which feed back and regulate distinct features of CLK/CYC function. Microarray studies have identified numerous rhythmically expressed transcripts, some of which are potential direct CLK targets. Here we demonstrate a circadian function for one such target, a bHLH-Orange repressor, CG17100/CLOCKWORK ORANGE (CWO). cwo is rhythmically expressed, and levels are reduced in Clk mutants, suggesting that cwo is CLK activated in vivo. cwo mutants display reduced-amplitude molecular and behavioral rhythms with lengthened periods. Molecular analysis suggests that CWO acts, in part, by repressing CLK target genes. We propose that CWO acts as a transcriptional and behavioral rhythm amplifier.
Figures


Similar articles
-
CLOCKWORK ORANGE promotes CLOCK-CYCLE activation via the putative Drosophila ortholog of CLOCK INTERACTING PROTEIN CIRCADIAN.Curr Biol. 2021 Oct 11;31(19):4207-4218.e4. doi: 10.1016/j.cub.2021.07.017. Epub 2021 Jul 30. Curr Biol. 2021. PMID: 34331859 Free PMC article.
-
The clockwork orange Drosophila protein functions as both an activator and a repressor of clock gene expression.J Biol Rhythms. 2008 Apr;23(2):103-16. doi: 10.1177/0748730407313817. J Biol Rhythms. 2008. PMID: 18375860
-
Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component.Genes Dev. 2007 Jul 1;21(13):1675-86. doi: 10.1101/gad.1552607. Epub 2007 Jun 19. Genes Dev. 2007. PMID: 17578907 Free PMC article.
-
Transcriptional feedback loop regulation, function, and ontogeny in Drosophila.Cold Spring Harb Symp Quant Biol. 2007;72:437-44. doi: 10.1101/sqb.2007.72.009. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419302 Free PMC article. Review.
-
The genetic and molecular regulation of sleep: from fruit flies to humans.Nat Rev Neurosci. 2009 Aug;10(8):549-60. doi: 10.1038/nrn2683. Nat Rev Neurosci. 2009. PMID: 19617891 Free PMC article. Review.
Cited by
-
The Catalytic and Non-catalytic Functions of the Brahma Chromatin-Remodeling Protein Collaborate to Fine-Tune Circadian Transcription in Drosophila.PLoS Genet. 2015 Jul 1;11(7):e1005307. doi: 10.1371/journal.pgen.1005307. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26132408 Free PMC article.
-
Genome-wide discovery of the daily transcriptome, DNA regulatory elements and transcription factor occupancy in the monarch butterfly brain.PLoS Genet. 2019 Jul 23;15(7):e1008265. doi: 10.1371/journal.pgen.1008265. eCollection 2019 Jul. PLoS Genet. 2019. PMID: 31335862 Free PMC article.
-
Model and Non-model Insects in Chronobiology.Front Behav Neurosci. 2020 Nov 26;14:601676. doi: 10.3389/fnbeh.2020.601676. eCollection 2020. Front Behav Neurosci. 2020. PMID: 33328925 Free PMC article. Review.
-
Microarray analysis of natural socially regulated plasticity in circadian rhythms of honey bees.J Biol Rhythms. 2012 Feb;27(1):12-24. doi: 10.1177/0748730411431404. J Biol Rhythms. 2012. PMID: 22306970 Free PMC article.
-
Circadian regulation of night feeding and daytime detoxification in a formidable Asian pest Spodoptera litura.Commun Biol. 2021 Mar 5;4(1):286. doi: 10.1038/s42003-021-01816-9. Commun Biol. 2021. PMID: 33674721 Free PMC article.
References
-
- Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–722. - PubMed
-
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. - PubMed
-
- Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277:14048–14052. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

