Roles of dynamic and reversible histone acetylation in plant development and polyploidy
- PMID: 17556080
- PMCID: PMC1950723
- DOI: 10.1016/j.bbaexp.2007.04.007
Roles of dynamic and reversible histone acetylation in plant development and polyploidy
Abstract
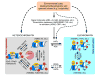
Transcriptional regulation in eukaryotes is not simply determined by the DNA sequence, but rather mediated through dynamic chromatin modifications and remodeling. Recent studies have shown that reversible and rapid changes in histone acetylation play an essential role in chromatin modification, induce genome-wide and specific changes in gene expression, and affect a variety of biological processes in response to internal and external signals, such as cell differentiation, growth, development, light, temperature, and abiotic and biotic stresses. Moreover, histone acetylation and deacetylation are associated with RNA interference and other chromatin modifications including DNA and histone methylation. The reversible changes in histone acetylation also contribute to cell cycle regulation and epigenetic silencing of rDNA and redundant genes in response to interspecific hybridization and polyploidy.
Figures

References
-
- Horn PJ, Peterson CL. Heterochromatin assembly: a new twist on an old model. Chromosome Res. 2006;14:83–94. - PubMed
-
- Kornberg RD. Structure of chromatin. Ann Rev Biochem. 1977;46:931–954. - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
-
- Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

