STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses
- PMID: 17556511
- PMCID: PMC2048787
- DOI: 10.1104/pp.107.099895
STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses
Abstract
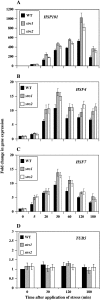
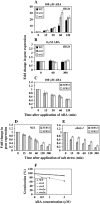
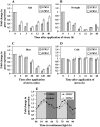
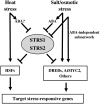
Two genes encoding Arabidopsis (Arabidopsis thaliana) DEAD-box RNA helicases were identified in a functional genomics screen as being down-regulated by multiple abiotic stresses. Mutations in either gene caused increased tolerance to salt, osmotic, and heat stresses, suggesting that the helicases suppress responses to abiotic stress. The genes were therefore designated STRESS RESPONSE SUPPRESSOR1 (STRS1; At1g31970) and STRS2 (At5g08620). In the strs mutants, salt, osmotic, and cold stresses induced enhanced expression of genes encoding the transcriptional activators DREB1A/CBF3 and DREB2A and a downstream DREB target gene, RD29A. Under heat stress, the strs mutants exhibited enhanced expression of the heat shock transcription factor genes, HSF4 and HSF7, and the downstream gene HEAT SHOCK PROTEIN101. Germination of mutant seed was hyposensitive to the phytohormone abscisic acid (ABA), but mutants showed up-regulated expression of genes encoding ABA-dependent stress-responsive transcriptional activators and their downstream targets. In wild-type plants, STRS1 and STRS2 expression was rapidly down-regulated by salt, osmotic, and heat stress, but not cold stress. STRS expression was also reduced by ABA, but salt stress led to reduced STRS expression in both wild-type and ABA-deficient mutant plants. Taken together, our results suggest that STRS1 and STRS2 attenuate the expression of stress-responsive transcriptional activators and function in ABA-dependent and ABA-independent abiotic stress signaling networks.
Figures
Similar articles
-
The Arabidopsis STRESS RESPONSE SUPPRESSOR DEAD-box RNA helicases are nucleolar- and chromocenter-localized proteins that undergo stress-mediated relocalization and are involved in epigenetic gene silencing.Plant J. 2014 Jul;79(1):28-43. doi: 10.1111/tpj.12533. Epub 2014 May 27. Plant J. 2014. PMID: 24724701
-
Functional characterization of DEAD-box RNA helicases in Arabidopsis thaliana under abiotic stress conditions.Plant Cell Physiol. 2008 Oct;49(10):1563-71. doi: 10.1093/pcp/pcn125. Epub 2008 Aug 24. Plant Cell Physiol. 2008. PMID: 18725370
-
DEAD-box RNA helicases and epigenetic control of abiotic stress-responsive gene expression.Plant Signal Behav. 2014;9(12):e977729. doi: 10.4161/15592324.2014.977729. Plant Signal Behav. 2014. PMID: 25517295 Free PMC article.
-
ABA-dependent and ABA-independent signaling in response to osmotic stress in plants.Curr Opin Plant Biol. 2014 Oct;21:133-139. doi: 10.1016/j.pbi.2014.07.009. Epub 2014 Aug 9. Curr Opin Plant Biol. 2014. PMID: 25104049 Review.
-
Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress.J Exp Bot. 2013 Jan;64(2):445-58. doi: 10.1093/jxb/ers354. J Exp Bot. 2013. PMID: 23307915 Review.
Cited by
-
Overexpression of Solanum habrochaites microRNA319d (sha-miR319d) confers chilling and heat stress tolerance in tomato (S. lycopersicum).BMC Plant Biol. 2019 May 23;19(1):214. doi: 10.1186/s12870-019-1823-x. BMC Plant Biol. 2019. PMID: 31122194 Free PMC article.
-
DEAD-Box RNA Helicase Family in Physic Nut (Jatropha curcas L.): Structural Characterization and Response to Salinity.Plants (Basel). 2024 Mar 21;13(6):905. doi: 10.3390/plants13060905. Plants (Basel). 2024. PMID: 38592921 Free PMC article.
-
Comprehensive transcriptome analysis reveals novel genes involved in cardiac glycoside biosynthesis and mlncRNAs associated with secondary metabolism and stress response in Digitalis purpurea.BMC Genomics. 2012 Jan 10;13:15. doi: 10.1186/1471-2164-13-15. BMC Genomics. 2012. PMID: 22233149 Free PMC article.
-
Recruitment of Arabidopsis RNA Helicase AtRH9 to the Viral Replication Complex by Viral Replicase to Promote Turnip Mosaic Virus Replication.Sci Rep. 2016 Jul 26;6:30297. doi: 10.1038/srep30297. Sci Rep. 2016. PMID: 27456972 Free PMC article.
-
Abscisic Acid and Abiotic Stress Tolerance in Crop Plants.Front Plant Sci. 2016 May 4;7:571. doi: 10.3389/fpls.2016.00571. eCollection 2016. Front Plant Sci. 2016. PMID: 27200044 Free PMC article. Review.
References
-
- Albrecht V, Weinl S, Blazevic D, D'Angelo C, Batistic O, Kolukisaoglu U, Bock R, Schulz B, Harter K, Kudla J (2003) The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J 36 457–470 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

