Exclusion of Na+ via sodium ATPase (PpENA1) ensures normal growth of Physcomitrella patens under moderate salt stress
- PMID: 17556514
- PMCID: PMC1949878
- DOI: 10.1104/pp.106.094946
Exclusion of Na+ via sodium ATPase (PpENA1) ensures normal growth of Physcomitrella patens under moderate salt stress
Abstract
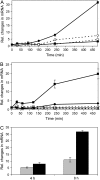
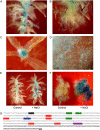
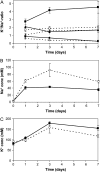
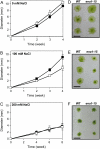
The bryophyte Physcomitrella patens is unlike any other plant identified to date in that it possesses a gene that encodes an ENA-type Na(+)-ATPase. To complement previous work in yeast (Saccharomyces cerevisiae), we determined the importance of having a Na(+)-ATPase in planta by conducting physiological analyses of PpENA1 in Physcomitrella. Expression studies showed that PpENA1 is up-regulated by NaCl and, to a lesser degree, by osmotic stress. Maximal induction is obtained after 8 h at 60 mm NaCl or above. No other abiotic stress tested led to significant increases in PpENA1 expression. In the gametophyte, strong expression was confined to the rhizoids, stem, and the basal part of the leaf. In the protonemata, expression was ubiquitous with a few filaments showing stronger expression. At 100 mm NaCl, wild-type plants were able to maintain a higher K(+)-to-Na(+) ratio than the PpENA1 (ena1) knockout gene, but at higher NaCl concentrations no difference was observed. Although no difference in chlorophyll content was observed between ena1 and wild type at 100 mm NaCl, the impaired Na(+) exclusion in ena1 plants led to an approximately 40% decrease in growth.
Figures

References
-
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285 1256–1258 - PubMed
-
- Ashton NW, Cove DJ (1977) Isolation and preliminary characterization of auxotrophic and analog resistant mutants of moss, Physcomitrella patens. Mol Gen Genet 154 87–95
-
- Ashton NW, Grimsley NH, Cove DJ (1979) Analysis of gametophytic development in the moss Physcomitrella patens using auxin and cytokinin resistant mutants. Planta 144 427–435 - PubMed
-
- Bañuelos MA, Quintero FJ, Rodríguez-Navarro A (1995) Functional expression of the ENA1(PMR2)-ATPase of Saccharomyces cerevisiae in Schizosaccharomyces pombe. Biochim Biophys Acta 1229 233–238 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

