Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families
- PMID: 17556529
- PMCID: PMC1899118
- DOI: 10.1101/gr.6214107
Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families
Abstract
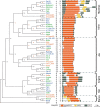
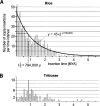
Although copia retrotransposons are major components of all plant genomes, the evolutionary relationships between individual copia families and between elements from different plant species are only poorly studied. We used 20 copia families from the large-genome plants barley and wheat to identify 46 families of homologous copia elements from rice and 22 from Arabidopsis, two plant species with much smaller genomes. In total, 599 copia elements were analyzed. Phylogenetic analysis showed that copia elements from the four species can be classified into six ancient lineages that existed before the divergence of monocots and dicots. The six lineages show a surprising degree of conservation in sequence organization and other characteristics across species. Additionally, the phylogenetic data suggest at least one case of horizontal gene transfer between the Arabidopsis and rice lineages. Insertion time estimates for 522 high-copy elements showed that retrotransposons from rice were active at different times in waves of activity lasting 0.5-2 million years, depending on the family, whereas elements from wheat and barley had longer periods of activity. We estimated that half of the rice copia elements are truncated or otherwise rearranged after approximately 790,000 yr, which is almost twice the half-life of Arabidopsis elements. In contrast, wheat and barley copia elements appear to have a massively longer half-life, beyond our ability to estimate from the available data. These findings suggest that genome size can be explained by the specific rate of DNA removal from the genome and the length of active periods of retrotransposon families.
Figures



References
-
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J.H., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J.H., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J.H., Zhang Z., Miller W., Lipman D.J., Zhang J.H., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. - PMC - PubMed
-
- ArabidopsisGenome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Bennett M.D., Smith J.B., Smith J.B. Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976;274:227–274. - PubMed
-
- Bossolini E., Wicker T., Knobel P., Keller B., Wicker T., Knobel P., Keller B., Knobel P., Keller B., Keller B. Comparison of orthologous loci from small grass genomes Brachypodium and rice: Implications for wheat genomics and grass genome annotation. Plant J. 2007;49:704–717. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
