Ionic effects on viral DNA packaging and portal motor function in bacteriophage phi 29
- PMID: 17556543
- PMCID: PMC2040884
- DOI: 10.1073/pnas.0701323104
Ionic effects on viral DNA packaging and portal motor function in bacteriophage phi 29
Abstract
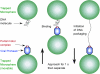
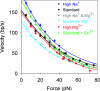
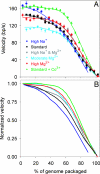
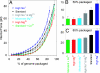
In many viruses, DNA is confined at such high density that its bending rigidity and electrostatic self-repulsion present a strong energy barrier in viral assembly. Therefore, a powerful molecular motor is needed to package the DNA into the viral capsid. Here, we investigate the role of electrostatic repulsion on single DNA packaging dynamics in bacteriophage phi 29 via optical tweezers measurements. We show that ionic screening strongly affects the packing forces, confirming the importance of electrostatic repulsion. Separately, we find that ions affect the motor function. We separate these effects through constant force measurements and velocity versus load measurements at both low and high capsid filling. Regarding motor function, we find that eliminating free Mg(2+) blocks initiation of packaging. In contrast, Na(+) is not required, but it increases the motor velocity by up to 50% at low load. Regarding internal resistance, we find that the internal force was lowest when Mg(2+) was the dominant ion or with the addition of 1 mM Co(3+). Forces resisting DNA confinement were up to approximately 80% higher with Na(+) as the dominant counterion, and only approximately 90% of the genome length could be packaged in this condition. The observed trend of the packing forces is in accord with that predicted by DNA charge-screening theory. However, the forces are up to six times higher than predicted by models that assume coaxial spooling of the DNA and interaction potentials derived from DNA condensation experiments. The forces are also severalfold higher than ejection forces measured with bacteriophage lambda.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
Stuffing a virus with DNA: dissecting viral genome packaging.Proc Natl Acad Sci U S A. 2007 Jul 3;104(27):11125-6. doi: 10.1073/pnas.0704764104. Epub 2007 Jun 26. Proc Natl Acad Sci U S A. 2007. PMID: 17595296 Free PMC article. No abstract available.
Similar articles
-
Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage phi29.Biophys J. 2008 Jan 1;94(1):159-67. doi: 10.1529/biophysj.107.104612. Epub 2007 Sep 7. Biophys J. 2008. PMID: 17827233 Free PMC article.
-
Experimental comparison of forces resisting viral DNA packaging and driving DNA ejection.Phys Rev E. 2017 May;95(5-1):052408. doi: 10.1103/PhysRevE.95.052408. Epub 2017 May 17. Phys Rev E. 2017. PMID: 28618627 Free PMC article.
-
The bacteriophage straight phi29 portal motor can package DNA against a large internal force.Nature. 2001 Oct 18;413(6857):748-52. doi: 10.1038/35099581. Nature. 2001. PMID: 11607035
-
Bacteriophage phi 29 DNA packaging.Adv Virus Res. 2002;58:255-94. doi: 10.1016/s0065-3527(02)58007-6. Adv Virus Res. 2002. PMID: 12205781 Review. No abstract available.
-
Effects of salts on internal DNA pressure and mechanical properties of phage capsids.J Mol Biol. 2011 Jan 7;405(1):18-23. doi: 10.1016/j.jmb.2010.10.039. Epub 2010 Oct 28. J Mol Biol. 2011. PMID: 21035458 Review.
Cited by
-
Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution.Nat Commun. 2015 Jul 6;6:7548. doi: 10.1038/ncomms8548. Nat Commun. 2015. PMID: 26144253 Free PMC article.
-
Understanding the physics of DNA using nanoscale single-molecule manipulation.Front Phys (Beijing). 2012 Oct;7(5):576-581. doi: 10.1007/s11467-012-0261-0. Front Phys (Beijing). 2012. PMID: 23467419 Free PMC article.
-
Three-way junction conformation dictates self-association of phage packaging RNAs.RNA Biol. 2016 Jul 2;13(7):635-45. doi: 10.1080/15476286.2016.1190075. Epub 2016 May 24. RNA Biol. 2016. PMID: 27217219 Free PMC article.
-
Osmotic pressure and packaging structure of caged DNA.Biophys J. 2008 Feb 1;94(3):737-46. doi: 10.1529/biophysj.107.112508. Epub 2007 Sep 21. Biophys J. 2008. PMID: 17890390 Free PMC article.
-
Systematic analysis of biological roles of charged amino acid residues located throughout the structured inner wall of a virus capsid.Sci Rep. 2018 Jun 22;8(1):9543. doi: 10.1038/s41598-018-27749-8. Sci Rep. 2018. PMID: 29934575 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

