Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism
- PMID: 17557806
- PMCID: PMC1955714
- DOI: 10.1105/tpc.107.051391
Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism
Abstract
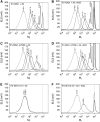
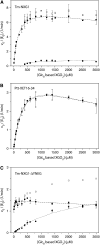
High-resolution, three-dimensional structures of the archetypal glycoside hydrolase family 16 (GH16) endo-xyloglucanases Tm-NXG1 and Tm-NXG2 from nasturtium (Tropaeolum majus) have been solved by x-ray crystallography. Key structural features that modulate the relative rates of substrate hydrolysis to transglycosylation in the GH16 xyloglucan-active enzymes were identified by structure-function studies of the recombinantly expressed enzymes in comparison with data for the strict xyloglucan endo-transglycosylase Ptt-XET16-34 from hybrid aspen (Populus tremula x Populus tremuloides). Production of the loop deletion variant Tm-NXG1-DeltaYNIIG yielded an enzyme that was structurally similar to Ptt-XET16-34 and had a greatly increased transglycosylation:hydrolysis ratio. Comprehensive bioinformatic analyses of XTH gene products, together with detailed kinetic data, strongly suggest that xyloglucanase activity has evolved as a gain of function in an ancestral GH16 XET to meet specific biological requirements during seed germination, fruit ripening, and rapid wall expansion.
Figures

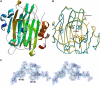

References
-
- Aubert, D., and Herzog, M. (1996). A new cDNA encoding a xyloglucan endo-transglycosylase-related polypeptide (AtXTR8) preferentially expressed in seedling, root and stem of Arabidopsis thaliana. Plant Sci. 121 187–196.
-
- Barbeyron, T., Gerard, A., Potin, P., Henrissat, B., and Kloareg, B. (1998). The kappa-carrageenase of the marine bacterium Cytophaga drobachiensis. Structural and phylogenetic relationships within family-16 glycoside hydrolases. Mol. Biol. Evol. 15 528–537. - PubMed
-
- Becnel, J., Natarajan, M., Kipp, A., and Braam, J. (2006). Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol. Biol. 61 451–467. - PubMed
-
- Bendtsen, J.D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340 783–795. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

