Synthesis and investigation of deoxyribonucleic acid/locked nucleic acid chimeric molecular beacons
- PMID: 17557813
- PMCID: PMC1919502
- DOI: 10.1093/nar/gkm358
Synthesis and investigation of deoxyribonucleic acid/locked nucleic acid chimeric molecular beacons
Abstract
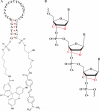
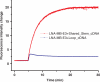
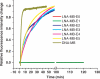
To take full advantage of locked nucleic acid (LNA) based molecular beacons (LNA-MBs) for a variety of applications including analysis of complex samples and intracellular monitoring, we have systematically synthesized a series of DNA/LNA chimeric MBs and studied the effect of DNA/LNA ratio in MBs on their thermodynamics, hybridization kinetics, protein binding affinity and enzymatic resistance. It was found that the LNA bases in a MB stem sequence had a significant effect on the stability of the hair-pin structure. The hybridization rates of LNA-MBs were significantly improved by lowering the DNA/LNA ratio in the probe, and most significantly, by having a shared-stem design for the LNA-MB to prevent sticky-end pairing. It was found that only MB sequences with DNA/LNA alternating bases or all LNA bases were able to resist nonspecific protein binding and DNase I digestion. Additional results showed that a sequence consisting of a DNA stretch less than three bases between LNA bases was able to block RNase H function. This study suggested that a shared-stem MB with a 4 base-pair stem and alternating DNA/LNA bases is desirable for intracellular applications as it ensures reasonable hybridization rates, reduces protein binding and resists nuclease degradation for both target and probes. These findings have implications on the design of LNA molecular probes for intracellular monitoring application, disease diagnosis and basic biological studies.
Figures




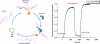



Similar articles
-
Locked nucleic acid molecular beacons.J Am Chem Soc. 2005 Nov 16;127(45):15664-5. doi: 10.1021/ja052498g. J Am Chem Soc. 2005. PMID: 16277483
-
Carba-LNA-5MeC/A/G/T modified oligos show nucleobase-specific modulation of 3'-exonuclease activity, thermodynamic stability, RNA selectivity, and RNase H elicitation: synthesis and biochemistry.J Org Chem. 2011 Jun 3;76(11):4408-31. doi: 10.1021/jo200073q. Epub 2011 May 4. J Org Chem. 2011. PMID: 21500818
-
Improved in situ hybridization efficiency with locked-nucleic-acid-incorporated DNA probes.Appl Environ Microbiol. 2006 Aug;72(8):5311-7. doi: 10.1128/AEM.03039-05. Appl Environ Microbiol. 2006. PMID: 16885281 Free PMC article.
-
LNA-antisense rivals siRNA for gene silencing.Curr Opin Drug Discov Devel. 2004 Mar;7(2):188-94. Curr Opin Drug Discov Devel. 2004. PMID: 15603252 Review.
-
Locked nucleic acids: a promising molecular family for gene-function analysis and antisense drug development.Curr Opin Mol Ther. 2001 Jun;3(3):239-43. Curr Opin Mol Ther. 2001. PMID: 11497347 Review.
Cited by
-
Nucleic acid beacons for long-term real-time intracellular monitoring.Anal Chem. 2008 Apr 15;80(8):3025-8. doi: 10.1021/ac702637w. Epub 2008 Mar 6. Anal Chem. 2008. PMID: 18321137 Free PMC article.
-
2'-O-Methyl molecular beacon: a promising molecular tool that permits elimination of sticky-end pairing and improvement of detection sensitivity.RSC Adv. 2020 Nov 14;10(68):41618-41624. doi: 10.1039/d0ra07341e. eCollection 2020 Nov 11. RSC Adv. 2020. PMID: 35516551 Free PMC article.
-
DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells.Proc Natl Acad Sci U S A. 2010 Jan 5;107(1):5-10. doi: 10.1073/pnas.0909611107. Epub 2009 Dec 22. Proc Natl Acad Sci U S A. 2010. PMID: 20080797 Free PMC article.
-
Molecular beacons: powerful tools for imaging RNA in living cells.J Nucleic Acids. 2011;2011:741723. doi: 10.4061/2011/741723. Epub 2011 Aug 22. J Nucleic Acids. 2011. PMID: 21876785 Free PMC article.
-
Recent Advances in the Molecular Beacon Technology for Live-Cell Single-Molecule Imaging.iScience. 2020 Nov 13;23(12):101801. doi: 10.1016/j.isci.2020.101801. eCollection 2020 Dec 18. iScience. 2020. PMID: 33299972 Free PMC article. Review.
References
-
- Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. - PubMed
-
- Tan WH, Wang KM, Drake TJ. Molecular beacons. Curr. Opin. Chem. Biol. 2004;8:547–553. - PubMed
-
- Li JWJ, Fang XH, Schuster SM, Tan WH. Molecular beacons: a novel approach to detect protein - DNA interactions. Angewandte Chemie-International Edition. 2000;39:1049–1052. - PubMed
-
- Tan WH, Fang XH, Li J, Liu XJ. Molecular beacons: a novel DNA probe for nucleic acid and protein studies. Chem. Eur. J. 2000;6:1107–1111. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

