Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain
- PMID: 17557820
- PMCID: PMC1952031
- DOI: 10.1128/JB.00655-07
Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain
Abstract
The cold shock response of Escherichia coli is elicited by downshift of temperature from 37 degrees C to 15 degrees C and is characterized by induction of several cold shock proteins, including CsdA, during the acclimation phase. CsdA, a DEAD-box protein, has been proposed to participate in a variety of processes, such as ribosome biogenesis, mRNA decay, translation initiation, and gene regulation. It is not clear which of the functions of CsdA play a role in its essential cold shock function or whether all do, and so far no protein has been shown to complement its function in vivo. Our screening of an E. coli genomic library for an in vivo counterpart of CsdA that can compensate for its absence at low temperature revealed only one protein, RhlE, another DEAD-box RNA helicase. We also observed that although not detected in our genetic screening, two cold shock-inducible proteins, namely, CspA, an RNA chaperone, and RNase R, an exonuclease, can also complement the cold shock function of CsdA. Interestingly, the absence of CsdA and RNase R leads to increased sensitivity of the cells to even moderate temperature downshifts. The correlation between the helicase activity of CsdA and the stability of mRNAs of cold-inducible genes was shown using cspA mRNA, which was significantly stabilized in the DeltacsdA cells, an effect counteracted by overexpression of wild-type CsdA or RNase R but not by that of the helicase-deficient mutant of CsdA. These results suggest that the primary role of CsdA in cold acclimation of cells is in mRNA decay and that its helicase activity is pivotal for promoting degradation of mRNAs stabilized at low temperature.
Figures

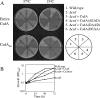



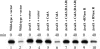
References
-
- Bizebard, T., I. Ferlenghi, I. Iost, and M. Dreyfus. 2004. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry 43:7857-7866. - PubMed
-
- Dammel, C. S., and H. F. Noller. 1995. Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev. 9:626-637. - PubMed
-
- Dersch, P., S. Kneip, and E. Bremer. 1994. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to a cold environment. Mol. Gen. Genet. 245:255-259. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

