Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae
- PMID: 17558514
- PMCID: PMC2084462
- DOI: 10.1007/s00412-007-0111-y
Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae
Abstract
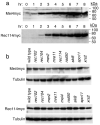
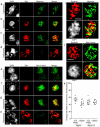
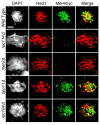
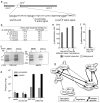
In most sexually reproducing organisms, meiotic recombination is initiated by DNA double-strand breaks (DSBs) formed by the Spo11 protein. In budding yeast, nine other proteins are also required for DSB formation, but the roles of these proteins and the interactions among them are poorly understood. We report further studies of the behaviors of these proteins. Consistent with other studies, we find that Mei4 and Rec114 bind to chromosomes from leptonema through early pachynema. Both proteins showed only limited colocalization with the meiotic cohesin subunit Rec8, suggesting that Mei4 and Rec114 associated preferentially with chromatin loops. Rec114 localization was independent of other DSB factors, but Mei4 localization was strongly dependent on Rec114 and Mer2. Systematic deletion analysis identified protein regions important for a previously described two-hybrid interaction between Mei4 and Rec114. We also report functional characterization of a previously misannotated 5' coding exon of REC102. Sequences encoded in this exon are essential for DSB formation and for Rec102 interaction with Rec104, Spo11, Rec114, and Mei4. Finally, we also examined genetic requirements for a set of previously described two-hybrid interactions that can be detected only when the reporter strain is induced to enter meiosis. This analysis reveals new functional dependencies for interactions among the DSB proteins. Taken together, these studies support the view that Mei4, Rec114, and Mer2 make up a functional subgroup that is distinct from other subgroups of the DSB proteins: Spo11-Ski8, Rec102-Rec104, and Mre11-Rad50-Xrs2. These studies also suggest that an essential function of Rec102 and Rec104 is to connect Mei4 and Rec114 to Spo11.
Figures


Similar articles
-
Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice.Genes Dev. 2010 Jun 15;24(12):1266-80. doi: 10.1101/gad.571710. Genes Dev. 2010. PMID: 20551173 Free PMC article.
-
Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation.Genetics. 2006 Aug;173(4):1969-81. doi: 10.1534/genetics.106.058768. Epub 2006 Jun 18. Genetics. 2006. PMID: 16783010 Free PMC article.
-
Genetic Interactions of Histone Modification Machinery Set1 and PAF1C with the Recombination Complex Rec114-Mer2-Mei4 in the Formation of Meiotic DNA Double-Strand Breaks.Int J Mol Sci. 2020 Apr 12;21(8):2679. doi: 10.3390/ijms21082679. Int J Mol Sci. 2020. PMID: 32290544 Free PMC article.
-
The multiple roles of the Mre11 complex for meiotic recombination.Chromosome Res. 2007;15(5):551-63. doi: 10.1007/s10577-007-1147-9. Chromosome Res. 2007. PMID: 17674145 Review.
-
Modulating and targeting meiotic double-strand breaks in Saccharomyces cerevisiae.Methods Mol Biol. 2009;557:27-33. doi: 10.1007/978-1-59745-527-5_3. Methods Mol Biol. 2009. PMID: 19799174 Review.
Cited by
-
Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice.Genes Dev. 2010 Jun 15;24(12):1266-80. doi: 10.1101/gad.571710. Genes Dev. 2010. PMID: 20551173 Free PMC article.
-
Genetics of mammalian meiosis: regulation, dynamics and impact on fertility.Nat Rev Genet. 2010 Feb;11(2):124-36. doi: 10.1038/nrg2723. Epub 2010 Jan 6. Nat Rev Genet. 2010. PMID: 20051984 Review.
-
Regulation Mechanisms of Meiotic Recombination Revealed from the Analysis of a Fission Yeast Recombination Hotspot ade6-M26.Biomolecules. 2022 Nov 26;12(12):1761. doi: 10.3390/biom12121761. Biomolecules. 2022. PMID: 36551189 Free PMC article. Review.
-
Regulating the formation of DNA double-strand breaks in meiosis.Genes Dev. 2008 Feb 1;22(3):286-92. doi: 10.1101/gad.1642308. Genes Dev. 2008. PMID: 18245442 Free PMC article. No abstract available.
-
Meiosis in budding yeast.Genetics. 2023 Oct 4;225(2):iyad125. doi: 10.1093/genetics/iyad125. Genetics. 2023. PMID: 37616582 Free PMC article.
References
-
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. - PubMed
-
- Arora C, Kee K, Maleki S, Keeney S. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol Cell. 2004;13:549–559. - PubMed
-
- Assenmacher N, Hopfner KP. MRE11/RAD50/NBS1: complex activities. Chromosoma. 2004;113:157–166. - PubMed
-
- Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

