CpG island mapping by epigenome prediction
- PMID: 17559301
- PMCID: PMC1892605
- DOI: 10.1371/journal.pcbi.0030110
CpG island mapping by epigenome prediction
Abstract
CpG islands were originally identified by epigenetic and functional properties, namely, absence of DNA methylation and frequent promoter association. However, this concept was quickly replaced by simple DNA sequence criteria, which allowed for genome-wide annotation of CpG islands in the absence of large-scale epigenetic datasets. Although widely used, the current CpG island criteria incur significant disadvantages: (1) reliance on arbitrary threshold parameters that bear little biological justification, (2) failure to account for widespread heterogeneity among CpG islands, and (3) apparent lack of specificity when applied to the human genome. This study is driven by the idea that a quantitative score of "CpG island strength" that incorporates epigenetic and functional aspects can help resolve these issues. We construct an epigenome prediction pipeline that links the DNA sequence of CpG islands to their epigenetic states, including DNA methylation, histone modifications, and chromatin accessibility. By training support vector machines on epigenetic data for CpG islands on human Chromosomes 21 and 22, we identify informative DNA attributes that correlate with open versus compact chromatin structures. These DNA attributes are used to predict the epigenetic states of all CpG islands genome-wide. Combining predictions for multiple epigenetic features, we estimate the inherent CpG island strength for each CpG island in the human genome, i.e., its inherent tendency to exhibit an open and transcriptionally competent chromatin structure. We extensively validate our results on independent datasets, showing that the CpG island strength predictions are applicable and informative across different tissues and cell types, and we derive improved maps of predicted "bona fide" CpG islands. The mapping of CpG islands by epigenome prediction is conceptually superior to identifying CpG islands by widely used sequence criteria since it links CpG island detection to their characteristic epigenetic and functional states. And it is superior to purely experimental epigenome mapping for CpG island detection since it abstracts from specific properties that are limited to a single cell type or tissue. In addition, using computational epigenetics methods we could identify high correlation between the epigenome and characteristics of the DNA sequence, a finding which emphasizes the need for a better understanding of the mechanistic links between genome and epigenome.
Conflict of interest statement
Figures


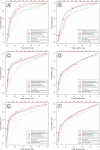
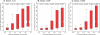


References
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
-
- Caiafa P, Zampieri M. DNA methylation and chromatin structure: The puzzling CpG islands. J Cell Biochem. 2005;94:257–265. - PubMed
-
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. - PubMed
-
- Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;14:R65–R76. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

