Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor
- PMID: 17559307
- PMCID: PMC1904358
- DOI: 10.1371/journal.pgen.0030094
Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor
Abstract
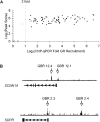
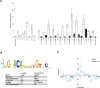
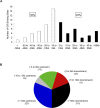
The glucocorticoid receptor (GR) associates with glucocorticoid response elements (GREs) and regulates selective gene transcription in a cell-specific manner. Native GREs are typically thought to be composite elements that recruit GR as well as other regulatory factors into functional complexes. We assessed whether GR occupancy is commonly a limiting determinant of GRE function as well as the extent to which core GR binding sequences and GRE architecture are conserved at functional loci. We surveyed 100-kb regions surrounding each of 548 known or potentially glucocorticoid-responsive genes in A549 human lung cells for GR-occupied GREs. We found that GR was bound in A549 cells predominately near genes responsive to glucocorticoids in those cells and not at genes regulated by GR in other cells. The GREs were positionally conserved at each responsive gene but across the set of responsive genes were distributed equally upstream and downstream of the transcription start sites, with 63% of them >10 kb from those sites. Strikingly, although the core GR binding sequences across the set of GREs varied extensively around a consensus, the precise sequence at an individual GRE was conserved across four mammalian species. Similarly, sequences flanking the core GR binding sites also varied among GREs but were conserved at individual GREs. We conclude that GR occupancy is a primary determinant of glucocorticoid responsiveness in A549 cells and that core GR binding sequences as well as GRE architecture likely harbor gene-specific regulatory information.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
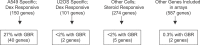

References
-
- Jobe AH, Ikegami M. Lung development and function in preterm infants in the surfactant treatment era. Annu Rev Physiol. 2000;62:825–846. - PubMed
-
- Yamamoto KR, Darimont BD, Wagner RL, Iniguez-Lluhi JA. Building transcriptional regulatory complexes: Signals and surfaces. Cold Spring Harb Symp Quant Biol. 1998;63:587–598. - PubMed
-
- Chandler VL, Maler BA, Yamamoto KR. DNA sequences bound specifically by glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell. 1983;33:489–499. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

