Essential and non-essential DNA replication genes in the model halophilic Archaeon, Halobacterium sp. NRC-1
- PMID: 17559652
- PMCID: PMC1906834
- DOI: 10.1186/1471-2156-8-31
Essential and non-essential DNA replication genes in the model halophilic Archaeon, Halobacterium sp. NRC-1
Abstract
Background: Information transfer systems in Archaea, including many components of the DNA replication machinery, are similar to those found in eukaryotes. Functional assignments of archaeal DNA replication genes have been primarily based upon sequence homology and biochemical studies of replisome components, but few genetic studies have been conducted thus far. We have developed a tractable genetic system for knockout analysis of genes in the model halophilic archaeon, Halobacterium sp. NRC-1, and used it to determine which DNA replication genes are essential.
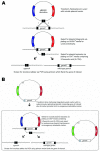
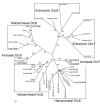
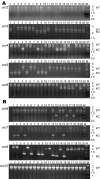
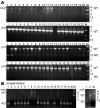
Results: Using a directed in-frame gene knockout method in Halobacterium sp. NRC-1, we examined nineteen genes predicted to be involved in DNA replication. Preliminary bioinformatic analysis of the large haloarchaeal Orc/Cdc6 family, related to eukaryotic Orc1 and Cdc6, showed five distinct clades of Orc/Cdc6 proteins conserved in all sequenced haloarchaea. Of ten orc/cdc6 genes in Halobacterium sp. NRC-1, only two were found to be essential, orc10, on the large chromosome, and orc2, on the minichromosome, pNRC200. Of the three replicative-type DNA polymerase genes, two were essential: the chromosomally encoded B family, polB1, and the chromosomally encoded euryarchaeal-specific D family, polD1/D2 (formerly called polA1/polA2 in the Halobacterium sp. NRC-1 genome sequence). The pNRC200-encoded B family polymerase, polB2, was non-essential. Accessory genes for DNA replication initiation and elongation factors, including the putative replicative helicase, mcm, the eukaryotic-type DNA primase, pri1/pri2, the DNA polymerase sliding clamp, pcn, and the flap endonuclease, rad2, were all essential. Targeted genes were classified as non-essential if knockouts were obtained and essential based on statistical analysis and/or by demonstrating the inability to isolate chromosomal knockouts except in the presence of a complementing plasmid copy of the gene.
Conclusion: The results showed that ten out of nineteen eukaryotic-type DNA replication genes are essential for Halobacterium sp. NRC-1, consistent with their requirement for DNA replication. The essential genes code for two of ten Orc/Cdc6 proteins, two out of three DNA polymerases, the MCM helicase, two DNA primase subunits, the DNA polymerase sliding clamp, and the flap endonuclease.
Figures


References
-
- Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, Swartzell S, Weir D, Hall J, Dahl TA, Welti R, Goo YA, Leithauser B, Keller K, Cruz R, Danson MJ, Hough DW, Maddocks DG, Jablonski PE, Krebs MP, Angevine CM, Dale H, Isenbarger TA, Peck RF, Pohlschroder M, Spudich JL, Jung KW, Alam M, Freitas T, Hou S, Daniels CJ, Dennis PP, Omer AD, Ebhardt H, Lowe TM, Liang P, Riley M, Hood L, DasSarma S. Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci U S A. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. - DOI - PMC - PubMed
-
- DasSarma S, Fleischmann EM. Halophiles. Plainview , NY: Cold Spring Harbor Laboratory Press; 1995.
-
- Berquist BR, Müller JA, DasSarma S. Genetic Systems for Halophilic Archaea. In: Oren A, Rainey F, editor. Methods in Microbiology. Vol. 35. Elsevier/Academic Press; 2005. pp. 649–680.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

