Can Clustal-style progressive pairwise alignment of multiple sequences be used in RNA secondary structure prediction?
- PMID: 17559658
- PMCID: PMC1904245
- DOI: 10.1186/1471-2105-8-190
Can Clustal-style progressive pairwise alignment of multiple sequences be used in RNA secondary structure prediction?
Abstract
Background: In ribonucleic acid (RNA) molecules whose function depends on their final, folded three-dimensional shape (such as those in ribosomes or spliceosome complexes), the secondary structure, defined by the set of internal basepair interactions, is more consistently conserved than the primary structure, defined by the sequence of nucleotides.
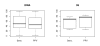
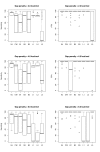
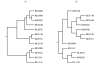
Results: The research presented here investigates the possibility of applying a progressive, pairwise approach to the alignment of multiple RNA sequences by simultaneously predicting an energy-optimized consensus secondary structure. We take an existing algorithm for finding the secondary structure common to two RNA sequences, Dynalign, and alter it to align profiles of multiple sequences. We then explore the relative successes of different approaches to designing the tree that will guide progressive alignments of sequence profiles to create a multiple alignment and prediction of conserved structure.
Conclusion: We have found that applying a progressive, pairwise approach to the alignment of multiple ribonucleic acid sequences produces highly reliable predictions of conserved basepairs, and we have shown how these predictions can be used as constraints to improve the results of a single-sequence structure prediction algorithm. However, we have also discovered that the amount of detail included in a consensus structure prediction is highly dependent on the order in which sequences are added to the alignment (the guide tree), and that if a consensus structure does not have sufficient detail, it is less likely to provide useful constraints for the single-sequence method.
Figures








References
-
- Sankoff D. Simultaneous solution of RNA folding, alignment and protosequence problems. SIAM J Appl Math. 1985;45:810–825. doi: 10.1137/0145048. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

