Recombinational micro-evolution of functionally different metallothionein promoter alleles from Orchesella cincta
- PMID: 17562010
- PMCID: PMC1913499
- DOI: 10.1186/1471-2148-7-88
Recombinational micro-evolution of functionally different metallothionein promoter alleles from Orchesella cincta
Abstract
Background: Metallothionein (mt) transcription is elevated in heavy metal tolerant field populations of Orchesella cincta (Collembola). This suggests that natural selection acts on transcriptional regulation of mt in springtails at sites where cadmium (Cd) levels in soil reach toxic values This study investigates the nature and the evolutionary origin of polymorphisms in the metallothionein promoter (pmt) and their functional significance for mt expression.
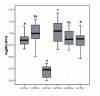
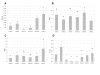
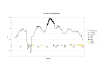
Results: We sequenced approximately 1600 bp upstream the mt coding region by genome walking. Nine pmt alleles were discovered in NW-European populations. They differ in the number of some indels, consensus transcription factor binding sites and core promoter elements. Extensive recombination events between some of the alleles can be inferred from the alignment. A deviation from neutral expectations was detected in a cadmium tolerant population, pointing towards balancing selection on some promoter stretches. Luciferase constructs were made from the most abundant alleles, and responses to Cd, paraquat (oxidative stress inducer) and moulting hormone were studied in cell lines. By using paraquat we were able to dissect the effect of oxidative stress from the Cd specific effect, and extensive differences in mt induction levels between these two stressors were observed.
Conclusion: The pmt alleles evolved by a number of recombination events, and exhibited differential inducibilities by Cd, paraquat and molting hormone. In a tolerant population from a metal contaminated site, promoter allele frequencies differed significantly from a reference site and nucleotide polymorphisms in some promoter stretches deviated from neutral expectations, revealing a signature of balancing selection. Our results suggest that the structural differences in the Orchesella cincta metallothionein promoter alleles contribute to the metallothionein -over-expresser phenotype in cadmium tolerant populations.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

