A complex and punctate distribution of three eukaryotic genes derived by lateral gene transfer
- PMID: 17562012
- PMCID: PMC1920508
- DOI: 10.1186/1471-2148-7-89
A complex and punctate distribution of three eukaryotic genes derived by lateral gene transfer
Abstract
Background: Lateral gene transfer is increasingly invoked to explain phylogenetic results that conflict with our understanding of organismal relationships. In eukaryotes, the most common observation interpreted in this way is the appearance of a bacterial gene (one that is not clearly derived from the mitochondrion or plastid) in a eukaryotic nuclear genome. Ideally such an observation would involve a single eukaryote or a small group of related eukaryotes encoding a gene from a specific bacterial lineage.
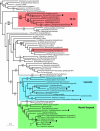
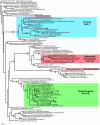
Results: Here we show that several apparently simple cases of lateral transfer are actually more complex than they originally appeared: in these instances we find that two or more distantly related eukaryotic groups share the same bacterial gene, resulting in a punctate distribution. Specifically, we describe phylogenies of three core carbon metabolic enzymes: transketolase, glyceraldehyde-3-phosphate dehydrogenase and ribulose-5-phosphate-3-epimerase. Phylogenetic trees of each of these enzymes includes a strongly-supported clade consisting of several eukaryotes that are distantly related at the organismal level, but whose enzymes are apparently all derived from the same lateral transfer. With less sampling any one of these examples would appear to be a simple case of bacterium-to-eukaryote lateral transfer; taken together, their evolutionary histories cannot be so simple. The distributions of these genes may represent ancient paralogy events or genes that have been transferred from bacteria to an ancient ancestor of the eukaryotes that retain them. They may alternatively have been transferred laterally from a bacterium to a single eukaryotic lineage and subsequently transferred between distantly related eukaryotes.
Conclusion: Determining how complex the distribution of a transferred gene is depends on the sampling available. These results show that seemingly simple cases may be revealed to be more complex with greater sampling, suggesting many bacterial genes found in eukaryotic genomes may have a punctate distribution.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials

