Large-scale identification of human genes implicated in epidermal barrier function
- PMID: 17562024
- PMCID: PMC2394760
- DOI: 10.1186/gb-2007-8-6-r107
Large-scale identification of human genes implicated in epidermal barrier function
Abstract
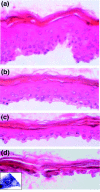
Background: During epidermal differentiation, keratinocytes progressing through the suprabasal layers undergo complex and tightly regulated biochemical modifications leading to cornification and desquamation. The last living cells, the granular keratinocytes (GKs), produce almost all of the proteins and lipids required for the protective barrier function before their programmed cell death gives rise to corneocytes. We present here the first analysis of the transcriptome of human GKs, purified from healthy epidermis by an original approach.
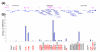
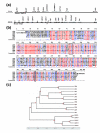
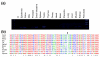
Results: Using the ORESTES method, 22,585 expressed sequence tags (ESTs) were produced that matched 3,387 genes. Despite normalization provided by this method (mean 4.6 ORESTES per gene), some highly transcribed genes, including that encoding dermokine, were overrepresented. About 330 expressed genes displayed less than 100 ESTs in UniGene clusters and are most likely to be specific for GKs and potentially involved in barrier function. This hypothesis was tested by comparing the relative expression of 73 genes in the basal and granular layers of epidermis by quantitative RT-PCR. Among these, 33 were identified as new, highly specific markers of GKs, including those encoding a protease, protease inhibitors and proteins involved in lipid metabolism and transport. We identified filaggrin 2 (also called ifapsoriasin), a poorly characterized member of the epidermal differentiation complex, as well as three new lipase genes clustered with paralogous genes on chromosome 10q23.31. A new gene of unknown function, C1orf81, is specifically disrupted in the human genome by a frameshift mutation.
Conclusion: These data increase the present knowledge of genes responsible for the formation of the skin barrier and suggest new candidates for genodermatoses of unknown origin.
Figures


References
-
- Lee WH, Jang S, Lee JS, Lee Y, Seo EY, You KH, Lee SC, Nam KI, Kim JM, Kee SH, et al. Molecular cloning and expression of human keratinocyte proline-rich protein (hKPRP), an epidermal marker isolated from calcium-induced differentiating keratinocytes. J Invest Dermatol. 2005;125:995–1000. doi: 10.1111/j.0022-202X.2005.23829.x. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

