Structure of synaptophysin: a hexameric MARVEL-domain channel protein
- PMID: 17562317
- PMCID: PMC1950735
- DOI: 10.1016/j.str.2007.04.011
Structure of synaptophysin: a hexameric MARVEL-domain channel protein
Abstract
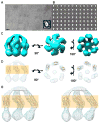
Synaptophysin I (SypI) is an archetypal member of the MARVEL-domain family of integral membrane proteins and one of the first synaptic vesicle proteins to be identified and cloned. Most all MARVEL-domain proteins are involved in membrane apposition and vesicle-trafficking events, but their precise role in these processes is unclear. We have purified mammalian SypI and determined its three-dimensional (3D) structure by using electron microscopy and single-particle 3D reconstruction. The hexameric structure resembles an open basket with a large pore and tenuous interactions within the cytosolic domain. The structure suggests a model for Synaptophysin's role in fusion and recycling that is regulated by known interactions with the SNARE machinery. This 3D structure of a MARVEL-domain protein provides a structural foundation for understanding the role of these important proteins in a variety of biological processes.
Figures


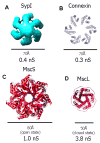

References
-
- Bai L, Spiwoks-Becker I, Leube RE. Transcriptome comparison of murine wild-type and synaptophysin-deficient retina reveals complete identity. Brain Res. 2006;1081:53–58. - PubMed
-
- Bass RB, Strop P, Barclay M, Rees DC. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science. 2002;298:1582–1587. - PubMed
-
- De Camilli P, Jahn R. Pathways to regulated exocytosis in neurons. Annu Rev Physiol. 1990;52:625–645. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

