Role of phospholipase Cgamma1 in cell spreading requires association with a beta-Pix/GIT1-containing complex, leading to activation of Cdc42 and Rac1
- PMID: 17562871
- PMCID: PMC1952113
- DOI: 10.1128/MCB.00778-07
Role of phospholipase Cgamma1 in cell spreading requires association with a beta-Pix/GIT1-containing complex, leading to activation of Cdc42 and Rac1
Abstract
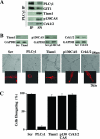
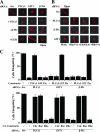
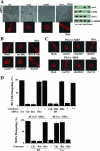
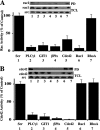
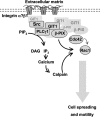
The significance of multiprotein signaling complexes in cell motility is becoming increasingly important. We have previously shown that phospholipase Cgamma1 (PLCgamma1) is critical for integrin-mediated cell spreading and motility (N. Jones et al., J. Cell Sci. 118:2695-2706, 2005). In the current study we show that, on a basement membrane-type matrix, PLCgamma1 associates with the adaptor protein GIT1 and the Rac1/Cdc42 guanine exchange factor beta-Pix; GIT1 and beta-Pix form tight complexes independently of PLCgamma1. The association of PLCgamma1 with the complex requires both GIT1 and beta-Pix and the specific array region (gammaSA) of PLCgamma1. Mutations of PLCgamma1 within the gammaSA region reveal that association with this complex is essential for the phosphorylation of PLCgamma1 and the progression to an elongated morphology after integrin engagement. Short interfering RNA (siRNA) depletion of either beta-Pix or GIT1 inhibited cell spreading in a fashion similar to that seen with siRNA against PLCgamma1. Furthermore, siRNA depletion of PLCgamma1, beta-Pix, or GIT1 inhibited Cdc42 and Rac1 activation, while constitutively active forms of Cdc42 or Rac1, but not RhoA, were able to rescue the elongation of these cells. Signaling of the PLCgamma1/GIT1/beta-Pix complex to Cdc42/Rac1 was found to involve the activation of calpains, calcium-dependent proteases. Therefore, we propose that the association of PLCgamma1 with complexes containing GIT1 and beta-Pix is essential for its role in integrin-mediated cell spreading and motility. As a component of this complex, PLCgamma1 is also involved in the activation of Cdc42 and Rac1.
Figures





Similar articles
-
β-PIX plays an important role in regulation of intestinal epithelial restitution by interacting with GIT1 and Rac1 after wounding.Am J Physiol Gastrointest Liver Physiol. 2018 Mar 1;314(3):G399-G407. doi: 10.1152/ajpgi.00296.2017. Epub 2017 Nov 30. Am J Physiol Gastrointest Liver Physiol. 2018. PMID: 29191942 Free PMC article.
-
The GIT/PIX complex: an oligomeric assembly of GIT family ARF GTPase-activating proteins and PIX family Rac1/Cdc42 guanine nucleotide exchange factors.Cell Signal. 2004 Sep;16(9):1001-11. doi: 10.1016/j.cellsig.2004.02.002. Cell Signal. 2004. PMID: 15212761
-
GIT1 activates p21-activated kinase through a mechanism independent of p21 binding.Mol Cell Biol. 2004 May;24(9):3849-59. doi: 10.1128/MCB.24.9.3849-3859.2004. Mol Cell Biol. 2004. PMID: 15082779 Free PMC article.
-
Expanding functions of GIT Arf GTPase-activating proteins, PIX Rho guanine nucleotide exchange factors and GIT-PIX complexes.J Cell Sci. 2016 May 15;129(10):1963-74. doi: 10.1242/jcs.179465. J Cell Sci. 2016. PMID: 27182061 Free PMC article. Review.
-
The guanine nucleotide exchange factor Tiam1: a Janus-faced molecule in cellular signaling.Cell Signal. 2014 Mar;26(3):483-91. doi: 10.1016/j.cellsig.2013.11.034. Epub 2013 Dec 2. Cell Signal. 2014. PMID: 24308970 Review.
Cited by
-
Finding the weakest link: exploring integrin-mediated mechanical molecular pathways.J Cell Sci. 2012 Jul 1;125(Pt 13):3025-38. doi: 10.1242/jcs.095794. Epub 2012 Jul 13. J Cell Sci. 2012. PMID: 22797926 Free PMC article. Review.
-
Inhibiting the Recruitment of PLCγ1 to Kaposi's Sarcoma Herpesvirus K15 Protein Reduces the Invasiveness and Angiogenesis of Infected Endothelial Cells.PLoS Pathog. 2015 Aug 21;11(8):e1005105. doi: 10.1371/journal.ppat.1005105. eCollection 2015 Aug. PLoS Pathog. 2015. PMID: 26295810 Free PMC article.
-
DDR1 promotes E-cadherin stability via inhibition of integrin-β1-Src activation-mediated E-cadherin endocytosis.Sci Rep. 2016 Nov 8;6:36336. doi: 10.1038/srep36336. Sci Rep. 2016. PMID: 27824116 Free PMC article.
-
Endogenous RhoG is dispensable for integrin-mediated cell spreading but contributes to Rac-independent migration.J Cell Sci. 2008 Jun 15;121(Pt 12):1981-9. doi: 10.1242/jcs.025130. Epub 2008 May 27. J Cell Sci. 2008. PMID: 18505794 Free PMC article.
-
Identification of components of the host type IA phosphoinositide 3-kinase pathway that promote internalization of Listeria monocytogenes.Infect Immun. 2012 Mar;80(3):1252-66. doi: 10.1128/IAI.06082-11. Epub 2011 Dec 12. Infect Immun. 2012. PMID: 22158742 Free PMC article.
References
-
- Bae, J. Y., S. J. Ahn, J. E. Lee, J. E. Kim, M. R. Han, W. Han, S. W. Kim, H. J. Shin, S. J. Lee, D. Park, and D. Y. Noh. 2005. BetaPix-a enhances the activity of phospholipase Cgamma1 by binding SH3 domain in breast cancer. J. Cell. Biochem. 94:1010-1016. - PubMed
-
- Baird, D., Q. Feng, and R. A. Cerione. 2005. The Cool-2/alpha-Pix protein mediates a Cdc42-Rac signaling cascade. Curr. Biol. 15:1-10. - PubMed
-
- Bialkowska, K., S. Kulkarni, X. Du, D. E. Goll, T. C. Saido, and J. E. Fox. 2000. Evidence that beta3 integrin-induced Rac activation involves the calpain-dependent formation of integrin clusters that are distinct from the focal complexes and focal adhesions that form as Rac and RhoA become active. J. Cell Biol. 151:685-696. - PMC - PubMed
-
- Carragher, N. O., S. M. Walker, L. A. Scott Carragher, F. Harris, T. K. Sawyer, V. G. Brunton, B. W. Ozanne, and M. C. Frame. 2006. Calpain 2 and Src dependence distinguishes mesenchymal and amoeboid modes of tumour cell invasion: a link to integrin function. Oncogene 25:5726-5740. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
