Still stratus not altocumulus: further evidence against the date/party hub distinction
- PMID: 17564494
- PMCID: PMC1892831
- DOI: 10.1371/journal.pbio.0050154
Still stratus not altocumulus: further evidence against the date/party hub distinction
Abstract
Analysis of multi-validated protein interaction data reveals networks with greater interconnectivity than the more segregated structures seen in previously available data. To help visualize this, the authors draw comparisons between continuous stratus clouds and altocumulus clouds.
Figures

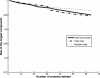

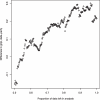
References
-
- Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. - PubMed
-
- Fraser HB. Modularity and evolutionary constraint on proteins. Nat Genet. 2005;37:351–352. - PubMed
-
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. - PubMed
-
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

