Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays
- PMID: 17564968
- PMCID: PMC1950910
- DOI: 10.1086/518809
Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays
Abstract
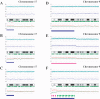
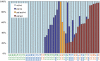
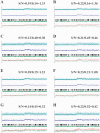
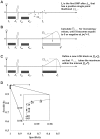
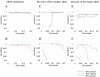
Loss of heterozygosity (LOH), either with or without accompanying copy-number loss, is a cardinal feature of cancer genomes that is tightly linked to cancer development. However, detection of LOH is frequently hampered by the presence of normal cell components within tumor specimens and the limitation in availability of constitutive DNA. Here, we describe a simple but highly sensitive method for genomewide detection of allelic composition, based on the Affymetrix single-nucleotide-polymorphism genotyping microarray platform, without dependence on the availability of constitutive DNA. By sensing subtle distortions in allele-specific signals caused by allelic imbalance with the use of anonymous controls, sensitive detection of LOH is enabled with accurate determination of allele-specific copy numbers, even in the presence of up to 70%-80% normal cell contamination. The performance of the new algorithm, called "AsCNAR" (allele-specific copy-number analysis using anonymous references), was demonstrated by detecting the copy-number neutral LOH, or uniparental disomy (UPD), in a large number of acute leukemia samples. We next applied this technique to detection of UPD involving the 9p arm in myeloproliferative disorders (MPDs), which is tightly associated with a homozygous JAK2 mutation. It revealed an unexpectedly high frequency of 9p UPD that otherwise would have been undetected and also disclosed the existence of multiple subpopulations having distinct 9p UPD within the same MPD specimen. In conclusion, AsCNAR should substantially improve our ability to dissect the complexity of cancer genomes and should contribute to our understanding of the genetic basis of human cancers.
Figures



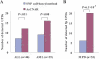

References
Web Resources
-
- BACPAC Resources Center, http://bacpac.chori.org/
-
- dChip, http://www.dchip.org/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for JAK2, AML, PV, ET, and IMF)
References
-
- Horvath A, Boikos S, Giatzakis C, Robinson-White A, Groussin L, Griffin KJ, Stein E, Levine E, Delimpasi G, Hsiao HP, et al (2006) A genome-wide scan identifies mutations in the gene encoding phosphodiesterase 11A4 (PDE11A) in individuals with adrenocortical hyperplasia. Nat Genet 38:794–800 10.1038/ng1809 - DOI - PubMed
-
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, et al (2005) Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365:1054–1061 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

