PrDOS: prediction of disordered protein regions from amino acid sequence
- PMID: 17567614
- PMCID: PMC1933209
- DOI: 10.1093/nar/gkm363
PrDOS: prediction of disordered protein regions from amino acid sequence
Abstract
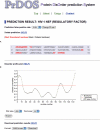
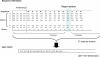
PrDOS is a server that predicts the disordered regions of a protein from its amino acid sequence (http://prdos.hgc.jp). The server accepts a single protein amino acid sequence, in either plain text or FASTA format. The prediction system is composed of two predictors: a predictor based on local amino acid sequence information and one based on template proteins. The server combines the results of the two predictors and returns a two-state prediction (order/disorder) and a disorder probability for each residue. The prediction results are sent by e-mail, and the server also provides a web-interface to check the results.
Figures
References
-
- Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:523–533. - PubMed
-
- Oldfield CJ, Ulrich EL, Cheng Y, Dunker AK, Markley JL. Addressing the intrinsic disorder bottleneck in structural proteomics. Proteins. 2005;59:444–453. - PubMed
-
- Fischer E. Einfluss der configuration auf die wirkung der enzyme. Ber. Dt. Chem. Ges. 1894;27:2985–2993.
-
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. - PubMed
-
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

