Hepatitis C virus nonstructural protein 5A modulates the toll-like receptor-MyD88-dependent signaling pathway in macrophage cell lines
- PMID: 17567694
- PMCID: PMC1951400
- DOI: 10.1128/JVI.00649-07
Hepatitis C virus nonstructural protein 5A modulates the toll-like receptor-MyD88-dependent signaling pathway in macrophage cell lines
Abstract
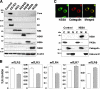
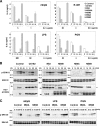
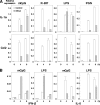
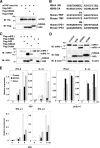
Hepatitis C virus (HCV) infection induces a wide range of chronic liver injuries; however, the mechanism through which HCV evades the immune surveillance system remains obscure. Blood dendritic cells (DCs) play a pivotal role in the recognition of viral infection and the induction of innate and adaptive immune responses. Several reports suggest that HCV infection induces the dysfunction of DCs in patients with chronic hepatitis C. Toll-like receptor (TLR) has been shown to play various roles in many viral infections; however, the involvement of HCV proteins in the TLR signaling pathway has not yet been precisely elucidated. In this study, we established mouse macrophage cell lines stably expressing HCV proteins and determined the effect of HCV proteins on the TLR signaling pathways. Immune cells expressing NS3, NS3/4A, NS4B, or NS5A were found to inhibit the activation of the TLR2, TLR4, TLR7, and TLR9 signaling pathways. Various genotypes of NS5A bound to MyD88, a major adaptor molecule in TLR, inhibited the recruitment of interleukin-1 receptor-associated kinase 1 to MyD88, and impaired cytokine production in response to TLR ligands. Amino acid residues 240 to 280, previously identified as the interferon sensitivity-determining region (ISDR) in NS5A, interacted with the death domain of MyD88, and the expression of a mutant NS5A lacking the ISDR partially restored cytokine production. These results suggest that the expression of HCV proteins modulates the TLR signaling pathway in immune cells.
Figures



References
-
- Aizaki, H., Y. Aoki, T. Harada, K. Ishii, T. Suzuki, S. Nagamori, G. Toda, Y. Matsuura, and T. Miyamura. 1998. Full-length complementary DNA of hepatitis C virus genome from an infectious blood sample. Hepatology 27:621-627. - PubMed
-
- Akira, S., and K. Takeda. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4:499-511. - PubMed
-
- Applequist, S. E., R. P. Wallin, and H. G. Ljunggren. 2002. Variable expression of Toll-like receptor in murine innate and adaptive immune cell lines. Int. Immunol. 14:1065-1074. - PubMed
-
- Bain, C., A. Fatmi, F. Zoulim, J. P. Zarski, C. Trepo, and G. Inchauspe. 2001. Impaired allostimulatory function of dendritic cells in chronic hepatitis C infection. Gastroenterology 120:512-524. - PubMed
-
- Bouffard, P., P. H. Hayashi, R. Acevedo, N. Levy, and J. B. Zeldis. 1992. Hepatitis C virus is detected in a monocyte/macrophage subpopulation of peripheral blood mononuclear cells of infected patients. J. Infect. Dis. 166:1276-1280. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

