Activation of cellular Arf GTPases by poliovirus protein 3CD correlates with virus replication
- PMID: 17567696
- PMCID: PMC1951455
- DOI: 10.1128/JVI.00840-07
Activation of cellular Arf GTPases by poliovirus protein 3CD correlates with virus replication
Abstract
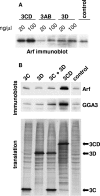
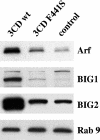
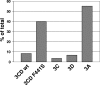
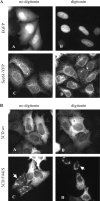
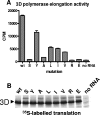
We have previously shown that synthesis of poliovirus protein 3CD in uninfected HeLa cell extracts induces an increased association with membranes of the cellular Arf GTPases, which are key players in cellular membrane traffic. Arfs cycle between an inactive, cytoplasmic, GDP-bound form and an active, membrane-associated, GTP-bound form. 3CD promotes binding of Arf to membranes by initiating recruitment to membranes of guanine nucleotide exchange factors (GEFs), BIG1 and BIG2. GEFs activate Arf by replacing GDP with GTP. In poliovirus-infected cells, there is a dramatic redistribution of cellular Arf pools that coincides with the reorganization of membranes used to form viral RNA replication complexes. Here we demonstrate that Arf translocation in vitro can be induced by purified recombinant 3CD protein; thus, concurrent translation of viral RNA is not required. Coexpression of 3C and 3D proteins was not sufficient to target Arf to membranes. 3CD expressed in HeLa cells was retained after treatment of the cells with digitonin, indicating that it may interact with a membrane-bound host factor. A F441S mutant of 3CD was shown previously to have lost Arf translocation activity and was also defective in attracting the corresponding GEFs to membranes. A series of other mutations were introduced at 3CD residue F441. Mutations that retained Arf translocation activity of 3CD also supported efficient growth of virus, regardless of their effects on 3D polymerase elongation activity. Those that abrogated Arf activation by 3CD generated quasi-infectious RNAs that produced some plaques from which revertants that always restored the Arf activation property of 3CD were rescued.
Figures

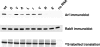
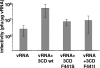
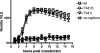
References
-
- Antonny, B., S. Beraud-Dufour, P. Chardin, and M. Chabre. 1997. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry 36:4675-4684. - PubMed
-
- Behnia, R., and S. Munro. 2005. Organelle identity and the signposts for membrane traffic. Nature 438:597-604. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

