o-Nitrotyrosine and p-iodophenylalanine as spectroscopic probes for structural characterization of SH3 complexes
- PMID: 17567746
- PMCID: PMC2206685
- DOI: 10.1110/ps.062726807
o-Nitrotyrosine and p-iodophenylalanine as spectroscopic probes for structural characterization of SH3 complexes
Abstract
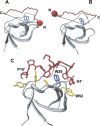
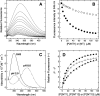
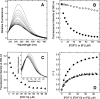
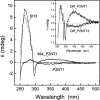
High-throughput screening of protein-protein and protein-peptide interactions is of high interest both for biotechnological and pharmacological applications. Here, we propose the use of the noncoded amino acids o-nitrotyrosine and p-iodophenylalanine as spectroscopic probes in combination with circular dichroism and fluorescence quenching techniques (i.e., collisional quenching and resonance energy transfer) as a means to determine the peptide orientation in complexes with SH3 domains. Proline-rich peptides bind SH3 modules in two alternative orientations, according to their sequence motifs, classified as class I and class II. The method was tested on an SH3 domain from a yeast myosin that is known to recognize specifically class I peptides. We exploited the fluorescence quenching effects induced by o-nitrotyrosine and p-iodophenylalanine on the fluorescence signal of a highly conserved Trp residue, which is the signature of SH3 domains and sits directly in the binding pocket. In particular, we studied how the introduction of the two probes at different positions of the peptide sequence (i.e., N-terminally or C-terminally) influences the spectroscopic properties of the complex. This approach provides clear-cut evidence of the orientation of the binding peptide in the SH3 pocket. The chemical strategy outlined here can be easily extended to other protein modules, known to bind linear sequence motifs in a highly directional manner.
Figures
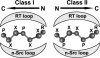
References
-
- Barlos K., Chatzi, O., Gatos, D., and Stavropoulos, G. 1991. 2-Chlorotrityl chloride resin. Studies on anchoring of Fmoc-amino acids and peptide cleavage. Int. J. Pept. Protein Res. 37: 513–520. - PubMed
-
- Berlman I.B. 1973. Empirical study of heavy-atom collisional quenching of the fluorescence state of aromatic compounds in solution. J. Phys. Chem. 77: 562–567.
-
- Carpino L.A. 1993. 1-Hydroxy-7-azabenzotriazole. An efficient peptide coupling additive. J. Am. Chem. Soc. 115: 4397–4398.
-
- Cesareni G., Panni, S., Nardelli, G., and Castagnoli, L. 2002. Can we infer peptide recognition specificity mediated by SH3 domains? FEBS Lett. 513: 38–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

